New bioinformatic tool for quick identification of functionally relevant endogenous retroviral inserts in human genome
- PMID: 25853282
- PMCID: PMC4612461
- DOI: 10.1080/15384101.2015.1022696
New bioinformatic tool for quick identification of functionally relevant endogenous retroviral inserts in human genome
Abstract
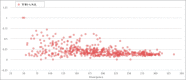
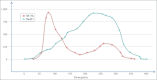
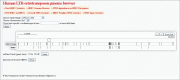
Endogenous retroviruses (ERVs) and LTR retrotransposons (LRs) occupy ∼8% of human genome. Deep sequencing technologies provide clues to understanding of functional relevance of individual ERVs/LRs by enabling direct identification of transcription factor binding sites (TFBS) and other landmarks of functional genomic elements. Here, we performed the genome-wide identification of human ERVs/LRs containing TFBS according to the ENCODE project. We created the first interactive ERV/LRs database that groups the individual inserts according to their familial nomenclature, number of mapped TFBS and divergence from their consensus sequence. Information on any particular element can be easily extracted by the user. We also created a genome browser tool, which enables quick mapping of any ERV/LR insert according to genomic coordinates, known human genes and TFBS. These tools can be used to easily explore functionally relevant individual ERV/LRs, and for studying their impact on the regulation of human genes. Overall, we identified ∼110,000 ERV/LR genomic elements having TFBS. We propose a hypothesis of "domestication" of ERV/LR TFBS by the genome milieu including subsequent stages of initial epigenetic repression, partial functional release, and further mutation-driven reshaping of TFBS in tight coevolution with the enclosing genomic loci.
Keywords: Database; LTR retrotransposon; endogenous retrovirus; genome browser; human genome; transcription factor binding site.
Figures



Similar articles
-
Long non-coding RNAs transcribed by ERV-9 LTR retrotransposon act in cis to modulate long-range LTR enhancer function.Nucleic Acids Res. 2017 May 5;45(8):4479-4492. doi: 10.1093/nar/gkx055. Nucleic Acids Res. 2017. PMID: 28132025 Free PMC article.
-
Profiling of Human Molecular Pathways Affected by Retrotransposons at the Level of Regulation by Transcription Factor Proteins.Front Immunol. 2018 Jan 30;9:30. doi: 10.3389/fimmu.2018.00030. eCollection 2018. Front Immunol. 2018. PMID: 29441061 Free PMC article.
-
ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):12565-12572. doi: 10.1073/pnas.1814589115. Epub 2018 Nov 19. Proc Natl Acad Sci U S A. 2018. PMID: 30455304 Free PMC article.
-
Utility of next-generation RNA-sequencing in identifying chimeric transcription involving human endogenous retroviruses.APMIS. 2016 Jan-Feb;124(1-2):127-39. doi: 10.1111/apm.12477. APMIS. 2016. PMID: 26818267 Review.
-
Nomenclature for endogenous retrovirus (ERV) loci.Retrovirology. 2018 Aug 28;15(1):59. doi: 10.1186/s12977-018-0442-1. Retrovirology. 2018. PMID: 30153831 Free PMC article. Review.
Cited by
-
HERVs and Cancer-A Comprehensive Review of the Relationship of Human Endogenous Retroviruses and Human Cancers.Biomedicines. 2023 Mar 17;11(3):936. doi: 10.3390/biomedicines11030936. Biomedicines. 2023. PMID: 36979914 Free PMC article. Review.
-
Living Organisms Author Their Read-Write Genomes in Evolution.Biology (Basel). 2017 Dec 6;6(4):42. doi: 10.3390/biology6040042. Biology (Basel). 2017. PMID: 29211049 Free PMC article. Review.
-
Friends-Enemies: Endogenous Retroviruses Are Major Transcriptional Regulators of Human DNA.Front Chem. 2017 Jun 8;5:35. doi: 10.3389/fchem.2017.00035. eCollection 2017. Front Chem. 2017. PMID: 28642863 Free PMC article. Review.
-
FNC: An Advanced Anticancer Therapeutic or Just an Underdog?Front Oncol. 2022 Feb 10;12:820647. doi: 10.3389/fonc.2022.820647. eCollection 2022. Front Oncol. 2022. PMID: 35223502 Free PMC article. Review.
-
Activation of human endogenous retroviruses and its physiological consequences.Nat Rev Mol Cell Biol. 2024 Mar;25(3):212-222. doi: 10.1038/s41580-023-00674-z. Epub 2023 Oct 23. Nat Rev Mol Cell Biol. 2024. PMID: 37872387 Review.
References
-
- Sverdlov ED. Retroviruses and primate evolution. Bioessays 2000; 2:161-71; http://dx.doi.org/10.1002/(SICI)1521-1878(200002)22:2%3c161::AID-BIES7%3... - DOI - PubMed
-
- Buzdin A. Human-specific endogenous retroviruses. Scientific World J 2007; 7:1848-68; PMID:18060323; http://dx.doi.org/10.1100/tsw.2007.270 - DOI - PMC - PubMed
-
- Hohn O, Hanke K, Bannert N. HERV-K(HML-2) the best preserved family of HERVs:Endogenization, expression, and implications in health and disease. Front Oncol 2013; 3:246; PMID:24066280; http://dx.doi.org/10.3389/fonc.2013.00246 - DOI - PMC - PubMed
-
- Dewannieux M, Heidmann T. Endogenous retroviruses:acquisition, amplification and taming of genome invaders. Curr Opin Virol 2013; 6:646-56; http://dx.doi.org/10.1016/j.coviro.2013.08.005 - DOI - PubMed
-
- Buzdin A, Ustyugova S, Khodosevich K, Mamedov I, Lebedev Y, Hunsmann G, Sverdlov E. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats:three master genes were active simultaneously during branching of hominoid lineages. Genomics 2003; 81(2):149-56; PMID:12620392; http://dx.doi.org/10.1016/S0888-7543(02)00027-7 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
