Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy
- PMID: 25853550
- PMCID: PMC4377852
- DOI: 10.1186/s13059-015-0620-6
Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy
Abstract
Background: While large-scale cancer genomic projects are comprehensively characterizing the mutational spectrum of various cancers, so far little attention has been devoted to either define the antigenicity of these mutations or to characterize the immune responses they elicit. Here we present a strategy to characterize the immunophenotypes and the antigen-ome of human colorectal cancer.
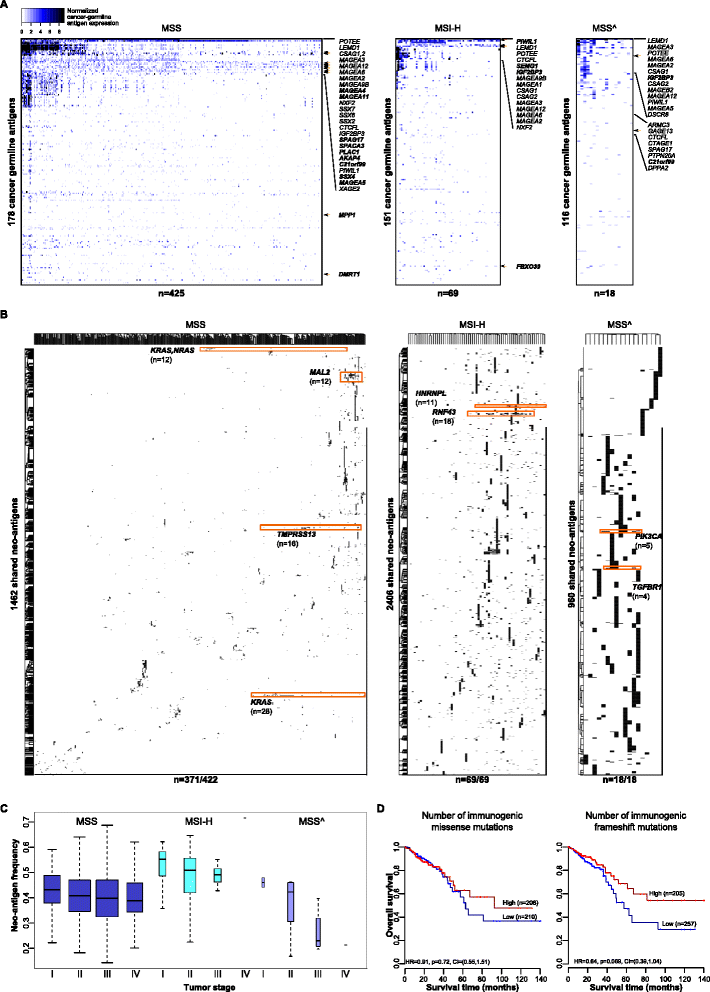
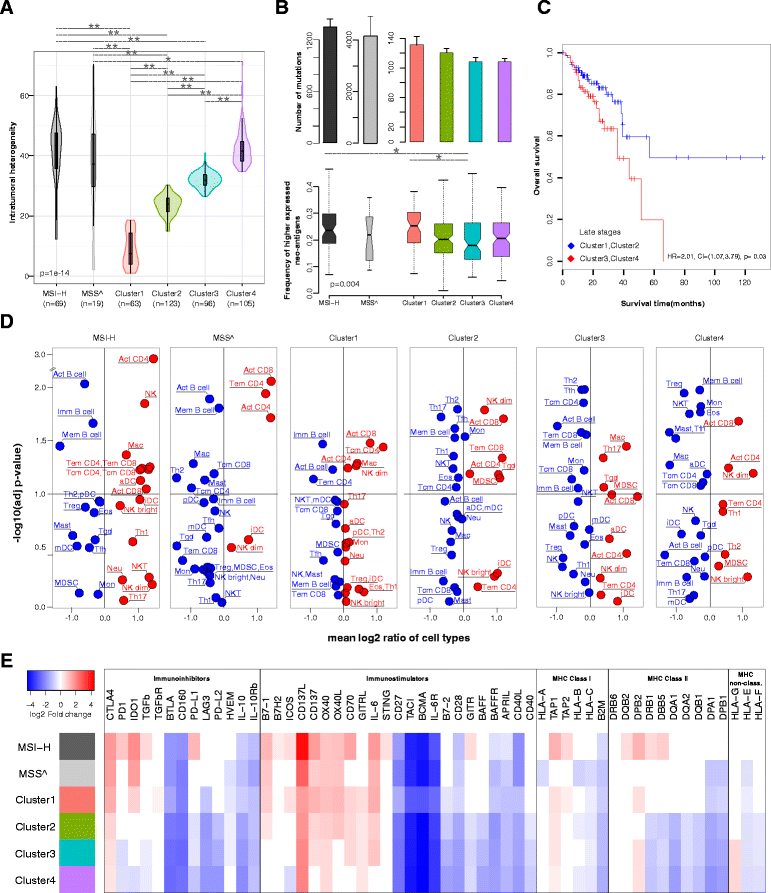
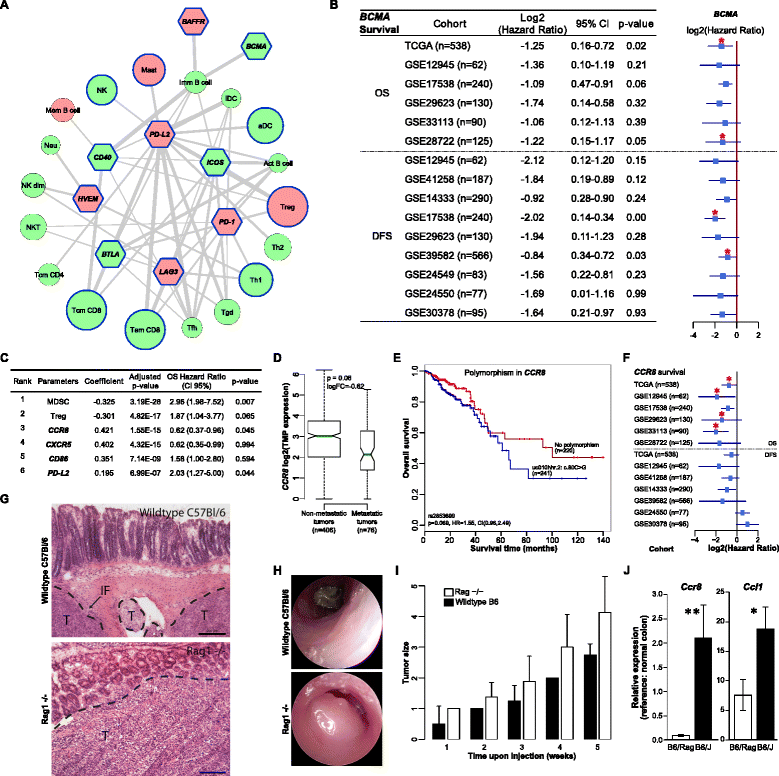
Results: We apply our strategy to a large colorectal cancer cohort (n = 598) and show that subpopulations of tumor-infiltrating lymphocytes are associated with distinct molecular phenotypes. The characterization of the antigenome shows that a large number of cancer-germline antigens are expressed in all patients. In contrast, neo-antigens are rarely shared between patients, indicating that cancer vaccination requires individualized strategy. Analysis of the genetic basis of the tumors reveals distinct tumor escape mechanisms for the patient subgroups. Hypermutated tumors are depleted of immunosuppressive cells and show upregulation of immunoinhibitory molecules. Non-hypermutated tumors are enriched with immunosuppressive cells, and the expression of immunoinhibitors and MHC molecules is downregulated. Reconstruction of the interaction network of tumor-infiltrating lymphocytes and immunomodulatory molecules followed by a validation with 11 independent cohorts (n = 1,945) identifies BCMA as a novel druggable target. Finally, linear regression modeling identifies major determinants of tumor immunogenicity, which include well-characterized modulators as well as a novel candidate, CCR8, which is then tested in an orthologous immunodeficient mouse model.
Conclusions: The immunophenotypes of the tumors and the cancer antigenome remain widely unexplored, and our findings represent a step toward the development of personalized cancer immunotherapies.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

