Engineering Escherichia coli into a protein delivery system for mammalian cells
- PMID: 25853840
- PMCID: PMC4487226
- DOI: 10.1021/acssynbio.5b00002
Engineering Escherichia coli into a protein delivery system for mammalian cells
Abstract
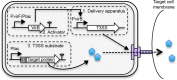
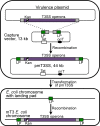
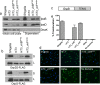
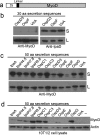
Many Gram-negative pathogens encode type 3 secretion systems, sophisticated nanomachines that deliver proteins directly into the cytoplasm of mammalian cells. These systems present attractive opportunities for therapeutic protein delivery applications; however, their utility has been limited by their inherent pathogenicity. Here, we report the reengineering of a laboratory strain of Escherichia coli with a tunable type 3 secretion system that can efficiently deliver heterologous proteins into mammalian cells, thereby circumventing the need for virulence attenuation. We first introduced a 31 kB region of Shigella flexneri DNA that encodes all of the information needed to form the secretion nanomachine onto a plasmid that can be directly propagated within E. coli or integrated into the E. coli chromosome. To provide flexible control over type 3 secretion and protein delivery, we generated plasmids expressing master regulators of the type 3 system from either constitutive or inducible promoters. We then constructed a Gateway-compatible plasmid library of type 3 secretion sequences to enable rapid screening and identification of sequences that do not perturb function when fused to heterologous protein substrates and optimized their delivery into mammalian cells. Combining these elements, we found that coordinated expression of the type 3 secretion system and modified target protein substrates produces a nonpathogenic strain that expresses, secretes, and delivers heterologous proteins into mammalian cells. This reengineered system thus provides a highly flexible protein delivery platform with potential for future therapeutic applications.
Keywords: bacterial engineering; protein delivery; synthetic biology; type 3 secretion system.
Figures
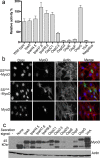
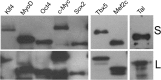
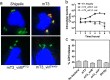
Similar articles
-
Rationale redesign of type III secretion systems: toward the development of non-pathogenic E. coli for in vivo delivery of therapeutic payloads.Curr Opin Microbiol. 2018 Feb;41:1-7. doi: 10.1016/j.mib.2017.10.011. Epub 2017 Nov 12. Curr Opin Microbiol. 2018. PMID: 29141238 Free PMC article. Review.
-
A Shigella flexneri virulence plasmid encoded factor controls production of outer membrane vesicles.G3 (Bethesda). 2014 Nov 5;4(12):2493-503. doi: 10.1534/g3.114.014381. G3 (Bethesda). 2014. PMID: 25378474 Free PMC article.
-
Identification of novel substrates of Shigella T3SA through analysis of its virulence plasmid-encoded secretome.PLoS One. 2017 Oct 26;12(10):e0186920. doi: 10.1371/journal.pone.0186920. eCollection 2017. PLoS One. 2017. PMID: 29073283 Free PMC article.
-
Functional cloning of Vibrio parahaemolyticus type III secretion system 1 in Escherichia coli K-12 strain as a molecular syringe.Biochem Biophys Res Commun. 2012 Oct 19;427(2):242-7. doi: 10.1016/j.bbrc.2012.09.018. Epub 2012 Sep 17. Biochem Biophys Res Commun. 2012. PMID: 22995311
-
Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors.FEMS Microbiol Lett. 2005 Nov 1;252(1):11-8. doi: 10.1016/j.femsle.2005.08.046. Epub 2005 Sep 15. FEMS Microbiol Lett. 2005. PMID: 16182469 Review.
Cited by
-
Heterologous Assembly of the Type VI Secretion System Empowers Laboratory Escherichia coli with Antimicrobial and Cell Penetration Capabilities.Appl Environ Microbiol. 2022 Oct 11;88(19):e0130522. doi: 10.1128/aem.01305-22. Epub 2022 Sep 26. Appl Environ Microbiol. 2022. PMID: 36154120 Free PMC article.
-
Rationale redesign of type III secretion systems: toward the development of non-pathogenic E. coli for in vivo delivery of therapeutic payloads.Curr Opin Microbiol. 2018 Feb;41:1-7. doi: 10.1016/j.mib.2017.10.011. Epub 2017 Nov 12. Curr Opin Microbiol. 2018. PMID: 29141238 Free PMC article. Review.
-
The type 3 secretion system requires actin polymerization to open translocon pores.PLoS Pathog. 2021 Sep 9;17(9):e1009932. doi: 10.1371/journal.ppat.1009932. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499700 Free PMC article.
-
pUdOs: Concise Plasmids for Bacterial and Mammalian Cells.ACS Synth Biol. 2024 Feb 16;13(2):485-497. doi: 10.1021/acssynbio.3c00408. Epub 2024 Jan 18. ACS Synth Biol. 2024. PMID: 38235654 Free PMC article.
-
PROT3EcT, engineered Escherichia coli for the targeted delivery of therapeutics.Trends Mol Med. 2023 Nov;29(11):968-969. doi: 10.1016/j.molmed.2023.07.007. Epub 2023 Aug 11. Trends Mol Med. 2023. PMID: 37574350 Free PMC article. No abstract available.
References
-
- Akeda Y.; Kimura T.; Yamasaki A.; Kodama T.; Iida T.; Honda T.; Oishi K. (2012) Functional cloning of Vibrio parahaemolyticus type III secretion system 1 in Escherichia coli K-12 strain as a molecular syringe. Biochem. Biophys. Res. Commun. 427, 242–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

