SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman
- PMID: 25853849
- PMCID: PMC4390220
- DOI: 10.1371/journal.pone.0123027
SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman
Abstract
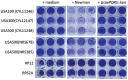
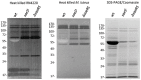
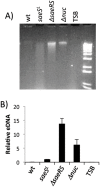
The SaeRS two-component regulatory system of Staphylococcus aureus is known to affect the expression of many genes. The SaeS protein is the histidine kinase responsible for phosphorylation of the response regulator SaeR. In S. aureus Newman, the sae system is constitutively expressed due to a point mutation in saeS, relative to other S. aureus strains, which results in substitution of proline for leucine at amino acid 18. Strain Newman is unable to form a robust biofilm and we report here that the biofilm-deficient phenotype is due to the saeSP allele. Replacement of the Newman saeSP with saeSL, or deletion of saeRS, resulted in a biofilm-proficient phenotype. Newman culture supernatants were observed to inhibit biofilm formation by other S. aureus strains, but did not affect biofilm formation by S. epidermidis. Culture supernatants of Newman saeSL or Newman ΔsaeRS had no significant effect on biofilm formation. The inhibitory factor was inactivated by incubation with proteinase K, but survived heating, indicating that the inhibitory protein is heat-stable. The inhibitory protein was found to affect the attachment step in biofilm formation, but had no effect on preformed biofilms. Replacement of saeSL with saeSP in the biofilm-proficient S. aureus USA300 FPR3757 resulted in the loss of biofilm formation. Culture supernatants of USA300 FPR3757 saeSP, did not inhibit biofilm formation by other staphylococci, suggesting that the inhibitory factor is produced but not secreted in the mutant strain. A number of biochemical methods were utilized to isolate the inhibitory protein. Although a number of candidate proteins were identified, none were found to be the actual inhibitor. In an effort to reduce the number of potential inhibitory genes, RNA-Seq analyses were done with wild-type strain Newman and the saeSL and ΔsaeRS mutants. RNA-Seq results indicated that sae regulates many genes that may affect biofilm formation by Newman.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P30GM103450/GM/NIGMS NIH HHS/United States
- AI113766/AI/NIAID NIH HHS/United States
- R01 AI113766/AI/NIAID NIH HHS/United States
- R01 AI074935/AI/NIAID NIH HHS/United States
- R56 AI074935/AI/NIAID NIH HHS/United States
- R01 AI087678/AI/NIAID NIH HHS/United States
- P20GM103429/GM/NIGMS NIH HHS/United States
- P30 GM103450/GM/NIGMS NIH HHS/United States
- R01 AI067857/AI/NIAID NIH HHS/United States
- P20 GM103625/GM/NIGMS NIH HHS/United States
- P20 GM103429/GM/NIGMS NIH HHS/United States
- P20GM103625/GM/NIGMS NIH HHS/United States
- R56 AI113766/AI/NIAID NIH HHS/United States
- AI074935/AI/NIAID NIH HHS/United States
- AI067857/AI/NIAID NIH HHS/United States
- AI087678/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

