Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging
- PMID: 25853883
- PMCID: PMC4375924
- DOI: 10.1186/s13059-015-0608-2
Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging
Abstract
Background: In studies of development and aging, the expression of many genes has been shown to undergo drastic changes at mRNA and protein levels. The connection between mRNA and protein expression level changes, as well as the role of posttranscriptional regulation in controlling expression level changes in postnatal development and aging, remains largely unexplored.
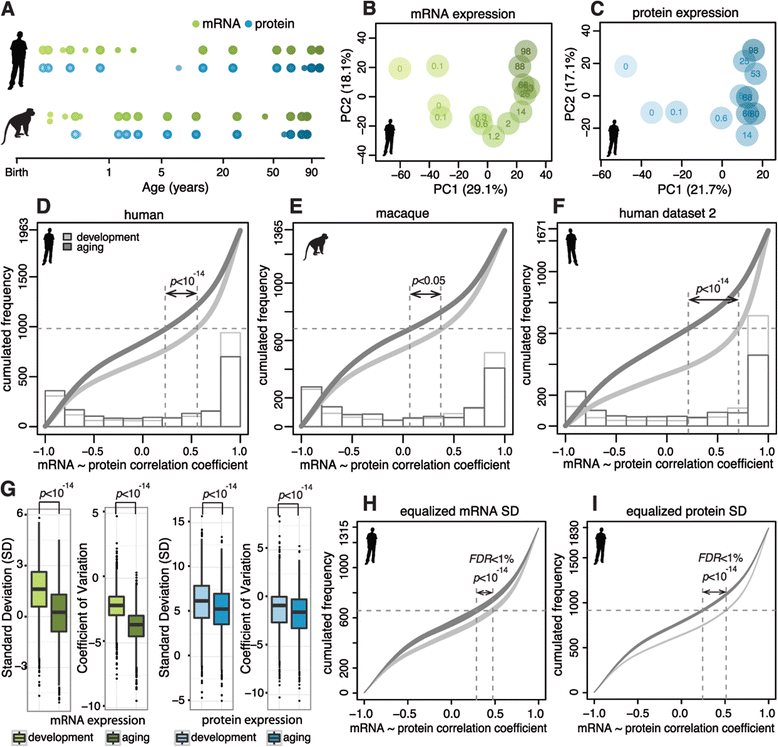
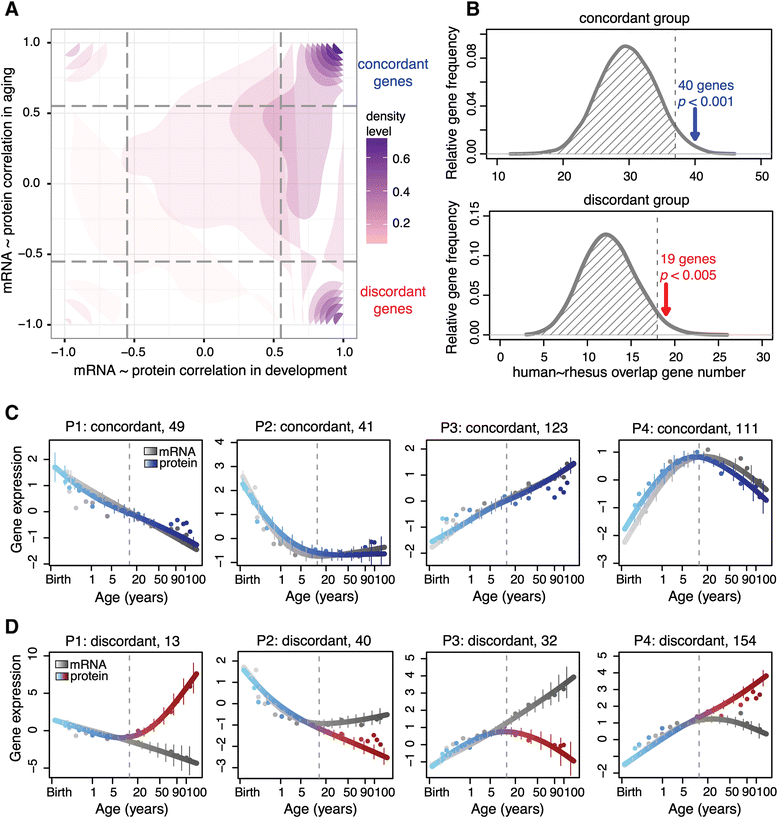
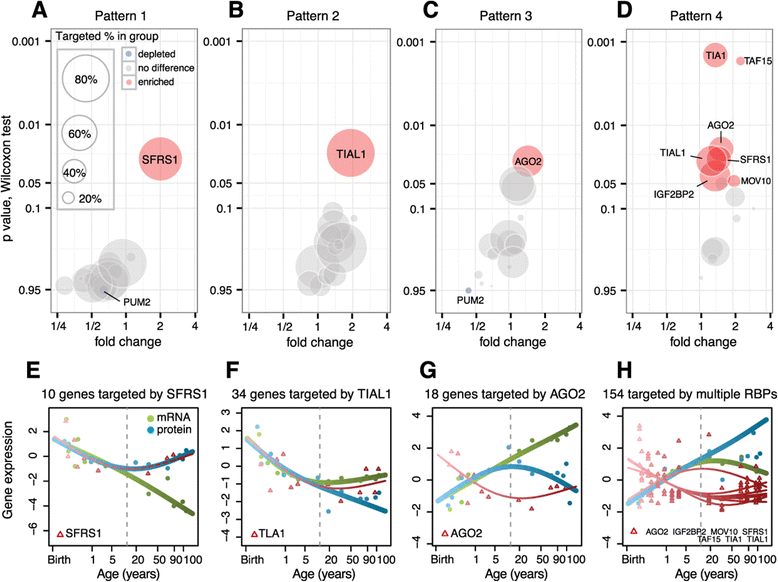
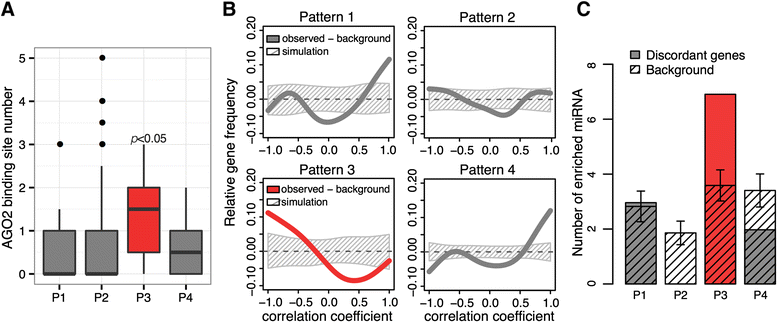
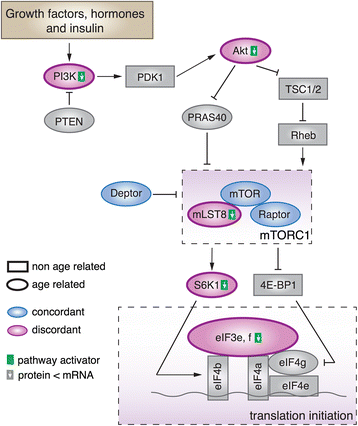
Results: Here, we survey mRNA and protein expression changes in the prefrontal cortex of humans and rhesus macaques over developmental and aging intervals of both species' lifespans. We find substantial decoupling of mRNA and protein expression levels in aging, but not in development. Genes showing increased mRNA/protein disparity in primate brain aging form expression patterns conserved between humans and macaques and are enriched in specific functions involving mammalian target of rapamycin (mTOR) signaling, mitochondrial function and neurodegeneration. Mechanistically, aging-dependent mRNA/protein expression decoupling could be linked to a specific set of RNA binding proteins and, to a lesser extent, to specific microRNAs.
Conclusions: Increased decoupling of mRNA and protein expression profiles observed in human and macaque brain aging results in specific co-expression profiles composed of genes with shared functions and shared regulatory signals linked to specific posttranscriptional regulators. Genes targeted and predicted to be targeted by the aging-dependent posttranscriptional regulation are associated with biological processes known to play important roles in aging and lifespan extension. These results indicate the potential importance of posttranscriptional regulation in modulating aging-dependent changes in humans and other species.
Figures
References
-
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56. - PubMed
-
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr Opin Genet Dev. 2011;21:491–7. - PubMed
-
- Janga SC, Vallabhaneni S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv Exp Med Biol. 2011;722:59–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

