MYB elongation is regulated by the nucleic acid binding of NFκB p50 to the intronic stem-loop region
- PMID: 25853889
- PMCID: PMC4390348
- DOI: 10.1371/journal.pone.0122919
MYB elongation is regulated by the nucleic acid binding of NFκB p50 to the intronic stem-loop region
Abstract
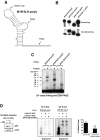
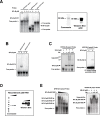
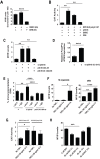
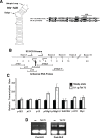
MYB transcriptional elongation is regulated by an attenuator sequence within intron 1 that has been proposed to encode a RNA stem loop (SLR) followed by a polyU tract. We report that NFκBp50 can bind the SLR polyU RNA and promote MYB transcriptional elongation together with NFκBp65. We identified a conserved lysine-rich motif within the Rel homology domain (RHD) of NFκBp50, mutation of which abrogated the interaction of NFκBp50 with the SLR polyU and impaired NFκBp50 mediated MYB elongation. We observed that the TAR RNA-binding region of Tat is homologous to the NFκBp50 RHD lysine-rich motif, a finding consistent with HIV Tat acting as an effector of MYB transcriptional elongation in an SLR dependent manner. Furthermore, we identify the DNA binding activity of NFκBp50 as a key component required for the SLR polyU mediated regulation of MYB. Collectively these results suggest that the MYB SLR polyU provides a platform for proteins to regulate MYB and reveals novel nucleic acid binding properties of NFκBp50 required for MYB regulation.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

