Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15
- PMID: 25854145
- PMCID: PMC4846425
- DOI: 10.1111/cbdd.12571
Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15
Abstract
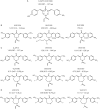
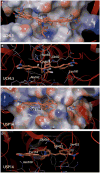
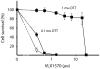
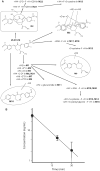
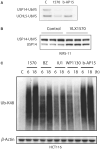
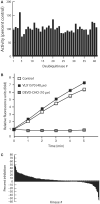
The ubiquitin-proteasome system (UPS) is increasingly recognized as a therapeutic target for the development of anticancer therapies. The success of the 20S proteasome core particle (20S CP) inhibitor bortezomib in the clinical management of multiple myeloma has raised the possibility of identifying other UPS components for therapeutic intervention. We previously identified the small molecule b-AP15 as an inhibitor of 19S proteasome deubiquitinase (DUB) activity. Building upon our previous data, we performed a structure-activity relationship (SAR) study on b-AP15 and identified VLX1570 as an analog with promising properties, including enhanced potency and improved solubility in aqueous solution. In silico modeling was consistent with interaction of VLX1570 with key cysteine residues located at the active sites of the proteasome DUBs USP14 and UCHL5. VLX1570 was found to inhibit proteasome deubiquitinase activity in vitro in a manner consistent with competitive inhibition. Furthermore, using active-site-directed probes, VLX1570 also inhibited proteasome DUB activity in exposed cells. Importantly, VLX1570 did not show inhibitory activity on a panel of recombinant non-proteasome DUBs, on recombinant kinases, or on caspase-3 activity, suggesting that VLX1570 is not an overtly reactive general enzyme inhibitor. Taken together, our data shows the chemical and biological properties of VLX1570 as an optimized proteasome DUB inhibitor.
Keywords: chalcone; deubiquitinase; in silico modeling; inhibitor; lead optimization; proteasome.
© 2015 John Wiley & Sons A/S.
Figures

Similar articles
-
The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells.Sci Rep. 2016 Jun 6;6:26979. doi: 10.1038/srep26979. Sci Rep. 2016. PMID: 27264969 Free PMC article.
-
Analysis of determinants for in vitro resistance to the small molecule deubiquitinase inhibitor b-AP15.PLoS One. 2019 Oct 22;14(10):e0223807. doi: 10.1371/journal.pone.0223807. eCollection 2019. PLoS One. 2019. PMID: 31639138 Free PMC article.
-
Targeted inhibition of the deubiquitinating enzymes, USP14 and UCHL5, induces proteotoxic stress and apoptosis in Waldenström macroglobulinaemia tumour cells.Br J Haematol. 2015 May;169(3):377-90. doi: 10.1111/bjh.13304. Epub 2015 Feb 17. Br J Haematol. 2015. PMID: 25691154 Free PMC article.
-
Proteasome deubiquitinases as novel targets for cancer therapy.Int J Biochem Cell Biol. 2012 Nov;44(11):1729-38. doi: 10.1016/j.biocel.2012.07.011. Epub 2012 Jul 20. Int J Biochem Cell Biol. 2012. PMID: 22819849 Review.
-
Inhibition of proteasome deubiquitinase activity: a strategy to overcome resistance to conventional proteasome inhibitors?Drug Resist Updat. 2015 Jul-Aug;21-22:20-9. doi: 10.1016/j.drup.2015.06.001. Epub 2015 Jul 6. Drug Resist Updat. 2015. PMID: 26183292 Review.
Cited by
-
Potential roles of UCH family deubiquitinases in tumorigenesis and chemical inhibitors developed against them.Am J Cancer Res. 2024 Jun 15;14(6):2666-2694. doi: 10.62347/OEGE2648. eCollection 2024. Am J Cancer Res. 2024. PMID: 39005671 Free PMC article. Review.
-
USP10 as a Potential Therapeutic Target in Human Cancers.Genes (Basel). 2022 May 6;13(5):831. doi: 10.3390/genes13050831. Genes (Basel). 2022. PMID: 35627217 Free PMC article. Review.
-
Drug resistance mechanisms and treatment strategies mediated by Ubiquitin-Specific Proteases (USPs) in cancers: new directions and therapeutic options.Mol Cancer. 2024 May 3;23(1):88. doi: 10.1186/s12943-024-02005-y. Mol Cancer. 2024. PMID: 38702734 Free PMC article. Review.
-
A novel deubiquitinase inhibitor b-AP15 triggers apoptosis in both androgen receptor-dependent and -independent prostate cancers.Oncotarget. 2017 Jun 28;8(38):63232-63246. doi: 10.18632/oncotarget.18774. eCollection 2017 Sep 8. Oncotarget. 2017. PMID: 28968984 Free PMC article.
-
Covalent Plasmodium falciparum-selective proteasome inhibitors exhibit a low propensity for generating resistance in vitro and synergize with multiple antimalarial agents.PLoS Pathog. 2019 Jun 6;15(6):e1007722. doi: 10.1371/journal.ppat.1007722. eCollection 2019 Jun. PLoS Pathog. 2019. PMID: 31170268 Free PMC article.
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. - PubMed
-
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. - PubMed
-
- D'Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. - PubMed
-
- D'Arcy P, Linder S. Molecular pathways: translational potential of deubiquitinases as drug targets. Clin Cancer Res. 2014;20:3908–3914. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

