Temperature-Sensitive Mutants in the Influenza A Virus RNA Polymerase: Alterations in the PA Linker Reduce Nuclear Targeting of the PB1-PA Dimer and Result in Viral Attenuation
- PMID: 25855727
- PMCID: PMC4474310
- DOI: 10.1128/JVI.00589-15
Temperature-Sensitive Mutants in the Influenza A Virus RNA Polymerase: Alterations in the PA Linker Reduce Nuclear Targeting of the PB1-PA Dimer and Result in Viral Attenuation
Abstract
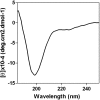
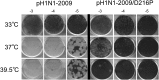
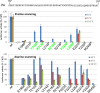
The influenza virus RNA-dependent RNA polymerase catalyzes genome replication and transcription within the cell nucleus. Efficient nuclear import and assembly of the polymerase subunits PB1, PB2, and PA are critical steps in the virus life cycle. We investigated the structure and function of the PA linker (residues 197 to 256), located between its N-terminal endonuclease domain and its C-terminal structured domain that binds PB1, the polymerase core. Circular dichroism experiments revealed that the PA linker by itself is structurally disordered. A large series of PA linker mutants exhibited a temperature-sensitive (ts) phenotype (reduced viral growth at 39.5°C versus 37°C/33°C), suggesting an alteration of folding kinetic parameters. The ts phenotype was associated with a reduced efficiency of replication/transcription of a pseudoviral reporter RNA in a minireplicon assay. Using a fluorescent-tagged PB1, we observed that ts and lethal PA mutants did not efficiently recruit PB1 to reach the nucleus at 39.5°C. A protein complementation assay using PA mutants, PB1, and β-importin IPO5 tagged with fragments of the Gaussia princeps luciferase showed that increasing the temperature negatively modulated the PA-PB1 and the PA-PB1-IPO5 interactions or complex stability. The selection of revertant viruses allowed the identification of different types of compensatory mutations located in one or the other of the three polymerase subunits. Two ts mutants were shown to be attenuated and able to induce antibodies in mice. Taken together, our results identify a PA domain critical for PB1-PA nuclear import and that is a "hot spot" to engineer ts mutants that could be used to design novel attenuated vaccines.
Importance: By targeting a discrete domain of the PA polymerase subunit of influenza virus, we were able to identify a series of 9 amino acid positions that are appropriate to engineer temperature-sensitive (ts) mutants. This is the first time that a large number of ts mutations were engineered in such a short domain, demonstrating that rational design of ts mutants can be achieved. We were able to associate this phenotype with a defect of transport of the PA-PB1 complex into the nucleus. Reversion substitutions restored the ability of the complex to move to the nucleus. Two of these ts mutants were shown to be attenuated and able to produce antibodies in mice. These results are of high interest for the design of novel attenuated vaccines and to develop new antiviral drugs.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Codon Deletions in the Influenza A Virus PA Gene Generate Temperature-Sensitive Viruses.J Virol. 2016 Jan 20;90(7):3684-93. doi: 10.1128/JVI.03101-15. J Virol. 2016. PMID: 26792748 Free PMC article.
-
Mapping the domain structure of the influenza A virus polymerase acidic protein (PA) and its interaction with the basic protein 1 (PB1) subunit.Virology. 2008 Sep 15;379(1):135-42. doi: 10.1016/j.virol.2008.06.022. Epub 2008 Jul 26. Virology. 2008. PMID: 18657841
-
A novel function of the N-terminal domain of PA in assembly of influenza A virus RNA polymerase.Biochem Biophys Res Commun. 2011 Nov 4;414(4):719-26. doi: 10.1016/j.bbrc.2011.09.142. Epub 2011 Oct 6. Biochem Biophys Res Commun. 2011. PMID: 22001919
-
Virulence of pandemic (H1N1) 2009 influenza A polymerase reassortant viruses.Virulence. 2011 Sep-Oct;2(5):422-6. doi: 10.4161/viru.2.5.17267. Epub 2011 Sep 1. Virulence. 2011. PMID: 21921678 Review.
-
Focusing on the Influenza Virus Polymerase Complex: Recent Progress in Drug Discovery and Assay Development.Curr Med Chem. 2019;26(13):2243-2263. doi: 10.2174/0929867325666180706112940. Curr Med Chem. 2019. PMID: 29984646 Free PMC article. Review.
Cited by
-
Naturally occurring PAE206K point mutation in 2009 H1N1 pandemic influenza viruses impairs viral replication at high temperatures.Virol Sin. 2024 Feb;39(1):71-80. doi: 10.1016/j.virs.2023.11.005. Epub 2023 Nov 16. Virol Sin. 2024. PMID: 37979619 Free PMC article.
-
The Influenza Virus Protein PB1-F2 Increases Viral Pathogenesis through Neutrophil Recruitment and NK Cells Inhibition.PLoS One. 2016 Oct 31;11(10):e0165361. doi: 10.1371/journal.pone.0165361. eCollection 2016. PLoS One. 2016. PMID: 27798704 Free PMC article.
-
The host RNA polymerase II C-terminal domain is the anchor for replication of the influenza virus genome.Nat Commun. 2024 Feb 5;15(1):1064. doi: 10.1038/s41467-024-45205-2. Nat Commun. 2024. PMID: 38316757 Free PMC article.
-
A novel E198K substitution in the PA gene of influenza A virus with reduced susceptibility to baloxavir acid.Arch Virol. 2022 Jul;167(7):1565-1570. doi: 10.1007/s00705-022-05456-0. Epub 2022 May 5. Arch Virol. 2022. PMID: 35511288 Free PMC article.
-
Genome-wide characterization of the seasonal H3N2 virus in Shanghai reveals natural temperature-sensitive strains conferred by the I668V mutation in the PA subunit.Emerg Microbes Infect. 2018 Oct 23;7(1):171. doi: 10.1038/s41426-018-0172-4. Emerg Microbes Infect. 2018. PMID: 30353004 Free PMC article.
References
-
- Shaw M, Palese P. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley P (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Honda A, Ishihama A. 1997. The molecular anatomy of influenza virus RNA polymerase. Biol Chem 378:483–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

