Inbreeding Affects Gene Expression Differently in Two Self-Incompatible Arabidopsis lyrata Populations with Similar Levels of Inbreeding Depression
- PMID: 25855783
- PMCID: PMC4833072
- DOI: 10.1093/molbev/msv086
Inbreeding Affects Gene Expression Differently in Two Self-Incompatible Arabidopsis lyrata Populations with Similar Levels of Inbreeding Depression
Abstract
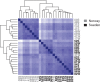
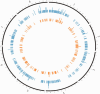
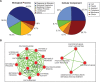
Knowledge of which genes and pathways are affected by inbreeding may help understanding the genetic basis of inbreeding depression, the potential for purging (selection against deleterious recessive alleles), and the transition from outcrossing to selfing. Arabidopsis lyrata is a predominantly self-incompatible perennial plant, closely related to the selfing model species A. thaliana. To examine how inbreeding affects gene expression, we compared the transcriptome of experimentally selfed and outcrossed A. lyrata originating from two Scandinavian populations that express similar inbreeding depression for fitness (∂ ≈ 0.80). The number of genes significantly differentially expressed between selfed and outcrossed individuals were 2.5 times higher in the Norwegian population (≈ 500 genes) than in the Swedish population (≈ 200 genes). In both populations, a majority of genes were upregulated on selfing (≈ 80%). Functional annotation analysis of the differentially expressed genes showed that selfed offspring were characterized by 1) upregulation of stress-related genes in both populations and 2) upregulation of photosynthesis-related genes in Sweden but downregulation in Norway. Moreover, we found that reproduction- and pollination-related genes were affected by inbreeding only in Norway. We conclude that inbreeding causes both general and population-specific effects. The observed common effects suggest that inbreeding generally upregulates rather than downregulates gene expression and affects genes associated with stress response and general metabolic activity. Population differences in the number of affected genes and in effects on the expression of photosynthesis-related genes show that the genetic basis of inbreeding depression can differ between populations with very similar levels of inbreeding depression.
Keywords: Arabidopsis lyrata; RNA-Seq; gene expression; inbreeding; stress-related genes.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Ayroles JF, Hughes KA, Rowe KC, Reedy MM, Rodriguez-Zas SL, Drnevich JM, Caceres CE, Paige KN. A genomewide assessment of inbreeding depression: gene number, function, and mode of action. Conserv Biol. 2009;23:920–930. - PubMed
-
- Barrett SCH, Charlesworth D. Effects of a change in the level of inbreeding on the genetic load. Nature. 1991;325:522–524. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

