SANSparallel: interactive homology search against Uniprot
- PMID: 25855811
- PMCID: PMC4489265
- DOI: 10.1093/nar/gkv317
SANSparallel: interactive homology search against Uniprot
Abstract
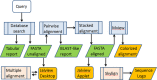
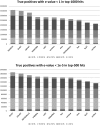
Proteins evolve by mutations and natural selection. The network of sequence similarities is a rich source for mining homologous relationships that inform on protein structure and function. There are many servers available to browse the network of homology relationships but one has to wait up to a minute for results. The SANSparallel webserver provides protein sequence database searches with immediate response and professional alignment visualization by third-party software. The output is a list, pairwise alignment or stacked alignment of sequence-similar proteins from Uniprot, UniRef90/50, Swissprot or Protein Data Bank. The stacked alignments are viewed in Jalview or as sequence logos. The database search uses the suffix array neighborhood search (SANS) method, which has been re-implemented as a client-server, improved and parallelized. The method is extremely fast and as sensitive as BLAST above 50% sequence identity. Benchmarks show that the method is highly competitive compared to previously published fast database search programs: UBLAST, DIAMOND, LAST, LAMBDA, RAPSEARCH2 and BLAT. The web server can be accessed interactively or programmatically at http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi. It can be used to make protein functional annotation pipelines more efficient, and it is useful in interactive exploration of the detailed evidence supporting the annotation of particular proteins of interest.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
NdPASA: a pairwise sequence alignment server for distantly related proteins.Bioinformatics. 2005 Oct 1;21(19):3803-5. doi: 10.1093/bioinformatics/bti619. Epub 2005 Aug 16. Bioinformatics. 2005. PMID: 16105904
-
Database similarity searches.Methods Mol Biol. 2008;484:361-78. doi: 10.1007/978-1-59745-398-1_24. Methods Mol Biol. 2008. PMID: 18592192
-
PANNZER2: a rapid functional annotation web server.Nucleic Acids Res. 2018 Jul 2;46(W1):W84-W88. doi: 10.1093/nar/gky350. Nucleic Acids Res. 2018. PMID: 29741643 Free PMC article.
-
Clustered sequence representation for fast homology search.J Comput Biol. 2007 Jun;14(5):594-614. doi: 10.1089/cmb.2007.R005. J Comput Biol. 2007. PMID: 17683263 Review.
-
NPS@: network protein sequence analysis.Trends Biochem Sci. 2000 Mar;25(3):147-50. doi: 10.1016/s0968-0004(99)01540-6. Trends Biochem Sci. 2000. PMID: 10694887 Review. No abstract available.
Cited by
-
DALI shines a light on remote homologs: One hundred discoveries.Protein Sci. 2023 Jan;32(1):e4519. doi: 10.1002/pro.4519. Protein Sci. 2023. PMID: 36419248 Free PMC article.
-
A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data.Comput Struct Biotechnol J. 2021 Nov 23;19:6301-6314. doi: 10.1016/j.csbj.2021.11.028. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34900140 Free PMC article. Review.
-
From reactants to products: computational methods for biosynthetic pathway design.Synth Syst Biotechnol. 2025 May 15;10(3):1038-1049. doi: 10.1016/j.synbio.2025.05.005. eCollection 2025 Sep. Synth Syst Biotechnol. 2025. PMID: 40510533 Free PMC article. Review.
-
Detection of novel tick-borne pathogen, Alongshan virus, in Ixodes ricinus ticks, south-eastern Finland, 2019.Euro Surveill. 2019 Jul;24(27):1900394. doi: 10.2807/1560-7917.ES.2019.24.27.1900394. Euro Surveill. 2019. PMID: 31290392 Free PMC article.
-
The transcriptome of Balamuthia mandrillaris trophozoites for structure-guided drug design.Sci Rep. 2021 Nov 4;11(1):21664. doi: 10.1038/s41598-021-99903-8. Sci Rep. 2021. PMID: 34737367 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

