WAXSiS: a web server for the calculation of SAXS/WAXS curves based on explicit-solvent molecular dynamics
- PMID: 25855813
- PMCID: PMC4489308
- DOI: 10.1093/nar/gkv309
WAXSiS: a web server for the calculation of SAXS/WAXS curves based on explicit-solvent molecular dynamics
Abstract
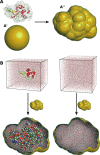
Small- and wide-angle X-ray scattering (SWAXS) has evolved into a powerful tool to study biological macromolecules in solution. The interpretation of SWAXS curves requires their accurate predictions from structural models. Such predictions are complicated by scattering contributions from the hydration layer and by effects from thermal fluctuations. Here, we describe the new web server WAXSiS (WAXS in solvent) that computes SWAXS curves based on explicit-solvent all-atom molecular dynamics (MD) simulations (http://waxsis.uni-goettingen.de/). The MD simulations provide a realistic model for both the hydration layer and the excluded solvent, thereby avoiding any solvent-related fitting parameters, while naturally accounting for thermal fluctuations.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Putnam C.D., Hammel M., Hura G.L., Tainer J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quart. Rev. Biophys. 2007;40:191–285. - PubMed
-
- Graewert M.A., Svergun D.I. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS) Curr. Opin. Struct. Biol. 2013;23:748–754. - PubMed
-
- Koch M.H., Vachette P., Svergun D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Quart. Rev. Biophys. 2003;36:147–227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

