Genetic diversity of five local Swedish chicken breeds detected by microsatellite markers
- PMID: 25855978
- PMCID: PMC4391840
- DOI: 10.1371/journal.pone.0120580
Genetic diversity of five local Swedish chicken breeds detected by microsatellite markers
Abstract
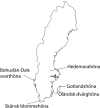
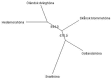
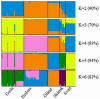
This study aimed at investigating the genetic diversity, relationship and population structure of 110 local Swedish chickens derived from five breeds (Gotlandshöna, Hedemorahöna, Öländsk dvärghöna, Skånsk blommehöna, and Bohuslän- Dals svarthöna, in the rest of the paper the shorter name Svarthöna is used) using 24 microsatellite markers. In total, one hundred thirteen alleles were detected in all populations, with a mean of 4.7 alleles per locus. For the five chicken breeds, the observed and expected heterozygosity ranged from 0.225 to 0.408 and from 0.231 to 0.515, with the lowest scores for the Svarthöna and the highest scores for the Skånsk blommehöna breeds, respectively. Similarly, the average within breed molecular kinship varied from 0.496 to 0.745, showing high coancestry, with Skånsk blommehöna having the lowest and Svarthöna the highest coancestry. Furthermore, all breeds showed significant deviations from Hardy-Weinberg expectations. Across the five breeds, the global heterozygosity deficit (FIT) was 0.545, population differentiation index (FST) was 0.440, and the global inbreeding of individuals within breed (FIS) was 0.187. The phylogenetic relationships of chickens were examined using neighbor-joining trees constructed at the level of breeds and individual samples. The neighbor-joining tree constructed at breed level revealed two main clusters, with Hedemorahöna and Öländsk dvärghöna breeds in one cluster, and Gotlandshöna and Svarthöna breeds in the second cluster leaving the Skånsk blommehöna in the middle. Based on the results of the STRUCTURE analysis, the most likely number of clustering of the five breeds was at K = 4, with Hedemorahöna, Gotlandshöna and Svarthöna breeds forming their own distinct clusters, while Öländsk dvärghöna and Skånsk blommehöna breeds clustered together. Losses in the overall genetic diversity of local Swedish chickens due to breeds extinction varied from -1.46% to -6.723%. The results of the current study can be used as baseline genetic information for genetic conservation program, for instance, to control inbreeding and to implement further genetic studies in local Swedish chickens.
Conflict of interest statement
Figures

References
-
- FAO (Food and Agriculture Organization). The State of the World’s Animal Genetic Resources for Food and Agriculture Edi: Barbara Rischkowsky & Dafydd Pilling. Rome. Italy; 2007. Available: http://www.fao.org/docrep/010/a1250e/a1250e00.htm
-
- Chen G, Bao W, Shu J, Ji C, Wang M, Eding H, et al. Assessment of Population Structure and Genetic Diversity of 15 Chinese Indigenous Chicken Breeds Using Microsatellite Markers. Asian-Aust J Anim Sci. 2008;21(3): 331–339.
-
- Granevitze Z, Hillel J, Chen GH, Cuc NTK, Feldman M, Eding H, et al. Genetic diversity within chicken populations from different continents and management histories. Anim Genet. 2007;38: 576–583. - PubMed
-
- FAO (Food and Agriculture Organization). Molecular genetic characterization of animal genetic resources FAO Animal Production and Health Guidelines. No. 9. Rome. Italy; 2011. Available: http://www.fao.org/docrep/014/i2413e/i2413e00.htm
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

