Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition
- PMID: 25856009
- PMCID: PMC7354103
- DOI: 10.1021/acs.jmedchem.5b00256
Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition
Abstract
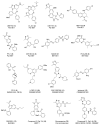
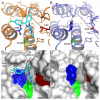
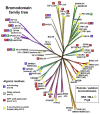
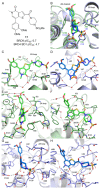
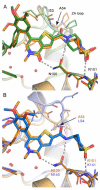
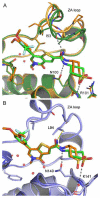
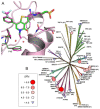
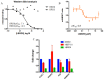
Acetylation of histone lysine residues is one of the most well-studied post-translational modifications of chromatin, selectively recognized by bromodomain "reader" modules. Inhibitors of the bromodomain and extra terminal domain (BET) family of bromodomains have shown profound anticancer and anti-inflammatory properties, generating much interest in targeting other bromodomain-containing proteins for disease treatment. Herein, we report the discovery of I-BRD9, the first selective cellular chemical probe for bromodomain-containing protein 9 (BRD9). I-BRD9 was identified through structure-based design, leading to greater than 700-fold selectivity over the BET family and 200-fold over the highly homologous bromodomain-containing protein 7 (BRD7). I-BRD9 was used to identify genes regulated by BRD9 in Kasumi-1 cells involved in oncology and immune response pathways and to the best of our knowledge, represents the first selective tool compound available to elucidate the cellular phenotype of BRD9 bromodomain inhibition.
Figures
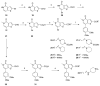
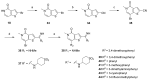
References
-
- Chung CW, Tough DF. Bromodomains: a new target class for small molecule drug discovery. Drug Discov Today: Ther Strategies. 2012;9:111–120.
-
- Chung CW. Small molecule bromodomain inhibitors: extending the druggable genome. Prog Med Chem. 2012;51:1–55. - PubMed
-
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases

