Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV)
- PMID: 25856093
- PMCID: PMC4391951
- DOI: 10.1371/journal.pone.0123126
Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV)
Abstract
The Middle East respiratory syndrome coronavirus (MERS-CoV), an emerging human coronavirus, causes severe acute respiratory illness with a 35% mortality rate. In light of the recent surge in reported infections we have developed asymmetric five-primer reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays for detection of MERS-CoV. Isothermal amplification assays will facilitate the development of portable point-of-care diagnostics that are crucial for management of emerging infections. The RT-LAMP assays are designed to amplify MERS-CoV genomic loci located within the open reading frame (ORF)1a and ORF1b genes and upstream of the E gene. Additionally we applied one-step strand displacement probes (OSD) for real-time sequence-specific verification of LAMP amplicons. Asymmetric amplification effected by incorporating a single loop primer in each assay accelerated the time-to-result of the OSD-RT-LAMP assays. The resulting assays could detect 0.02 to 0.2 plaque forming units (PFU) (5 to 50 PFU/ml) of MERS-CoV in infected cell culture supernatants within 30 to 50 min and did not cross-react with common human respiratory pathogens.
Conflict of interest statement
Figures
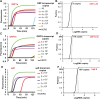
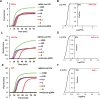
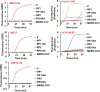
References
-
- Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39):pii = 20285. Epub 2012/10/09. PubMed . - PubMed
-
- Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382(9909):1993–2002. Epub 2013/09/24. 10.1016/S0140-6736(13)61887-5 PubMed . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

