Comparative genome analyses of Serratia marcescens FS14 reveals its high antagonistic potential
- PMID: 25856195
- PMCID: PMC4391916
- DOI: 10.1371/journal.pone.0123061
Comparative genome analyses of Serratia marcescens FS14 reveals its high antagonistic potential
Abstract
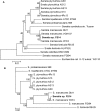
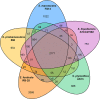
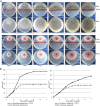
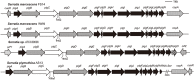
S. marcescens FS14 was isolated from an Atractylodes macrocephala Koidz plant that was infected by Fusarium oxysporum and showed symptoms of root rot. With the completion of the genome sequence of FS14, the first comprehensive comparative-genomic analysis of the Serratia genus was performed. Pan-genome and COG analyses showed that the majority of the conserved core genes are involved in basic cellular functions, while genomic factors such as prophages contribute considerably to genome diversity. Additionally, a Type I restriction-modification system, a Type III secretion system and tellurium resistance genes are found in only some Serratia species. Comparative analysis further identified that S. marcescens FS14 possesses multiple mechanisms for antagonism against other microorganisms, including the production of prodigiosin, bacteriocins, and multi-antibiotic resistant determinants as well as chitinases. The presence of two evolutionarily distinct Type VI secretion systems (T6SSs) in FS14 may provide further competitive advantages for FS14 against other microbes. To our knowledge, this is the first report of comparative analysis on T6SSs in the genus, which identifies four types of T6SSs in Serratia spp.. Competition bioassays of FS14 against the vital plant pathogenic bacterium Ralstonia solanacearum and fungi Fusarium oxysporum and Sclerotinia sclerotiorum were performed to support our genomic analyses, in which FS14 demonstrated high antagonistic activities against both bacterial and fungal phytopathogens.
Conflict of interest statement
Figures



References
-
- Zheng CC, Wang CL, Fu L, Wang WW, Xu DQ. Isolation and characterization of the Serratia sp. FS14 secreting thermostable DNase and protease.2011; 38: 228–236.
-
- Hejazi A, Falkiner FR. Serratia marcescens. J Med Microbiol. 1997; 46: 903–912. - PubMed
-
- De Vleesschauwer D, Höfte M. Using Serratia plymuthica to control fungal pathogens of plants. CAB Reviews. 2003; 2(46):189–257.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

