A genomic safe haven for mutant complementation in Cryptococcus neoformans
- PMID: 25856300
- PMCID: PMC4391909
- DOI: 10.1371/journal.pone.0122916
A genomic safe haven for mutant complementation in Cryptococcus neoformans
Abstract
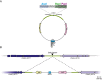
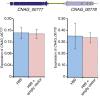
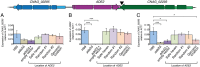
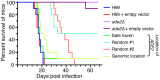
Just as Koch's postulates formed the foundation of early infectious disease study, Stanley Falkow's molecular Koch's postulates define best practice in determining whether a specific gene contributes to virulence of a pathogen. Fundamentally, these molecular postulates state that if a gene is involved in virulence, its removal will compromise virulence. Likewise, its reintroduction should restore virulence to the mutant. These approaches are widely employed in Cryptococcus neoformans, where gene deletion via biolistic transformation is a well-established technique. However, the complementation of these mutants is less straightforward. Currently, one of three approaches will be taken: the gene is reintroduced at the original locus, the gene is reintroduced into a random site in the genome, or the mutant is not complemented at all. Depending on which approach is utilized, the mutant may be complemented but other genes are potentially disrupted in the process. To counter the drawbacks of the current approaches to complementation we have created a new tool to assist in this key step in the study of a gene's role in virulence. We have identified and characterized a small gene-free region in the C. neoformans genome dubbed the "safe haven", and constructed a plasmid vector that targets DNA constructs to this preselected site. The plasmid vector integrates with high frequency, effectively complementing a mutant strain without disrupting adjacent genes. qRT-PCR of the flanking genes on either side of the safe haven site following integration of the targeting vector revealed no changes in their expression, and no secondary phenotypes were observed in a range of phenotypic assays including an intranasal murine infection model. Combined, these data confirm that we have successfully created a much-needed molecular resource for the Cryptococcus community, enabling the reliable fulfillment of the molecular Koch's postulates.
Conflict of interest statement
Figures




References
-
- Koch. 2 Die Aetiologie der Tuberkulose. 1884; 1–88 p.
-
- Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988; 10 Suppl 2: S274–276. - PubMed
-
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998; 14: 953–961. - PubMed
-
- Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology-Uk. 2000; 146: 1969–1975. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

