MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF
- PMID: 25857263
- PMCID: PMC4816102
- DOI: 10.1038/cdd.2015.30
MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF
Abstract
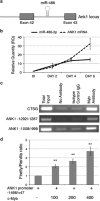
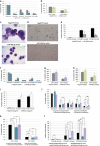
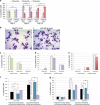
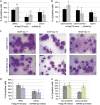
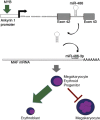
The transcription factor MYB has a key role in hematopoietic progenitor cells (HPCs) lineage choice, by enhancing erythropoiesis at the expense of megakaryopoiesis. We previously demonstrated that MYB controls erythroid versus megakaryocyte lineage decision by transactivating KLF1 and LMO2 expression. To further unravel the molecular mechanisms through which MYB affects lineage fate decision, we performed the integrative analysis of miRNA and mRNA changes in MYB-silenced human primary CD34+ HPCs. Among the miRNAs with the highest number of predicted targets, we focused our studies on hsa-miR-486-3p by demonstrating that MYB controls miR-486-3p expression through the transactivation of its host gene, ankyrin-1 (ANK1) and that miR-486-3p affects HPCs commitment. Indeed, overexpression and knockdown experiments demonstrated that miR-486-3p supports the erythropoiesis while restraining the megakaryopoiesis. Of note, miR-486-3p also favors granulocyte differentiation while repressing the macrophage differentiation. To shed some light on the molecular mechanisms through which miR-486-3p affects HPCs lineage commitment, we profiled the gene expression changes upon miR-486-3p overexpression in CD34+ cells. Among the genes downregulated in miR-486-3p-overexpressing HPCs and computationally predicted to be miR-486-3p targets, we identified MAF as a miR-486-3p target by 3'UTR luciferase reporter assay. Noteworthy, MAF overexpression was able to partially reverse the effects of miR-486-3p overexpression on erythroid versus megakaryocyte lineage choice. Moreover, the MYB/MAF co-silencing constrained the skewing of erythroid versus megakaryocyte lineage commitment in MYB-silenced CD34+ cells, by restraining the expansion of megakaryocyte lineage while partially rescuing the impairment of erythropoiesis. Therefore, our data collectively demonstrate that MYB favors erythropoiesis and restrains megakaryopoiesis through the transactivation of miR-486-3p expression and the subsequent downregulation of MAF. As a whole, our study uncovers the MYB/miR-486-3p/MAF axis as a new mechanism underlying the MYB-driven control of erythroid versus megakaryocyte lineage fate decision.
Figures


References
-
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991; 65: 677–689. - PubMed
-
- Bianchi E, Zini R, Salati S, Tenedini E, Norfo R, Tagliafico E et al. c-myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood 2010; 116: e99–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

