Controlled induction of DNA double-strand breaks in the mouse liver induces features of tissue ageing
- PMID: 25858675
- PMCID: PMC4394211
- DOI: 10.1038/ncomms7790
Controlled induction of DNA double-strand breaks in the mouse liver induces features of tissue ageing
Abstract
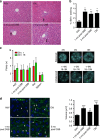
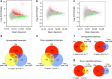
DNA damage has been implicated in ageing, but direct evidence for a causal relationship is lacking, owing to the difficulty of inducing defined DNA lesions in cells and tissues without simultaneously damaging other biomolecules and cellular structures. Here we directly test whether highly toxic DNA double-strand breaks (DSBs) alone can drive an ageing phenotype using an adenovirus-based system based on tetracycline-controlled expression of the SacI restriction enzyme. We deliver the adenovirus to mice and compare molecular and cellular end points in the liver with normally aged animals. Treated, 3-month-old mice display many, but not all signs of normal liver ageing as early as 1 month after treatment, including ageing pathologies, markers of senescence, fused mitochondria and alterations in gene expression profiles. These results, showing that DSBs alone can cause distinct ageing phenotypes in mouse liver, provide new insights in the role of DNA damage as a driver of tissue ageing.
Figures




References
-
- Jackson S. P. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696 (2002) . - PubMed
-
- Mine-Hattab J. & Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517 (2012) . - PubMed
-
- Soutoglou E. & Misteli T. On the contribution of spatial genome organization to cancerous chromosome translocations. J. Natl Cancer. Inst. Monogr. 39, 16–19 (2008) . - PubMed
-
- San Filippo J., Sung P. & Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008) . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

