Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9
- PMID: 25859012
- PMCID: PMC5007610
- DOI: 10.1093/hmg/ddv120
Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9
Abstract
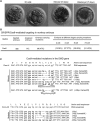
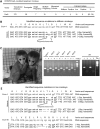
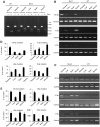
CRISPR/Cas9 has been used to genetically modify genomes in a variety of species, including non-human primates. Unfortunately, this new technology does cause mosaic mutations, and we do not yet know whether such mutations can functionally disrupt the targeted gene or cause the pathology seen in human disease. Addressing these issues is necessary if we are to generate large animal models of human diseases using CRISPR/Cas9. Here we used CRISPR/Cas9 to target the monkey dystrophin gene to create mutations that lead to Duchenne muscular dystrophy (DMD), a recessive X-linked form of muscular dystrophy. Examination of the relative targeting rate revealed that Crispr/Cas9 targeting could lead to mosaic mutations in up to 87% of the dystrophin alleles in monkey muscle. Moreover, CRISPR/Cas9 induced mutations in both male and female monkeys, with the markedly depleted dystrophin and muscle degeneration seen in early DMD. Our findings indicate that CRISPR/Cas9 can efficiently generate monkey models of human diseases, regardless of inheritance patterns. The presence of degenerated muscle cells in newborn Cas9-targeted monkeys suggests that therapeutic interventions at the early disease stage may be effective at alleviating the myopathy.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., et al. (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 156, 836–843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

