On the family-free DCJ distance and similarity
- PMID: 25859276
- PMCID: PMC4391664
- DOI: 10.1186/s13015-015-0041-9
On the family-free DCJ distance and similarity
Abstract
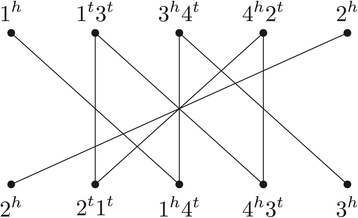
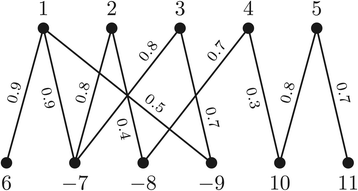
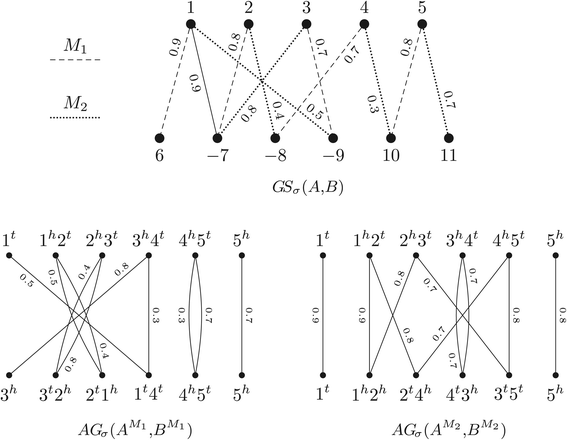
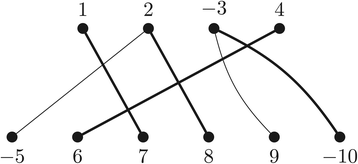
Structural variation in genomes can be revealed by many (dis)similarity measures. Rearrangement operations, such as the so called double-cut-and-join (DCJ), are large-scale mutations that can create complex changes and produce such variations in genomes. A basic task in comparative genomics is to find the rearrangement distance between two given genomes, i.e., the minimum number of rearragement operations that transform one given genome into another one. In a family-based setting, genes are grouped into gene families and efficient algorithms have already been presented to compute the DCJ distance between two given genomes. In this work we propose the problem of computing the DCJ distance of two given genomes without prior gene family assignment, directly using the pairwise similarities between genes. We prove that this new family-free DCJ distance problem is APX-hard and provide an integer linear program to its solution. We also study a family-free DCJ similarity and prove that its computation is NP-hard.
Keywords: DCJ; Family-free genome comparison; Genome rearrangement.
Figures
References
-
- Sankoff D. Proc. of CPM 1992. LNCS, vol. 644. Heidelberg: Springer Verlag; 1992. Edit distance for genome comparison based on non-local operations.
-
- Bergeron A, Mixtacki J, Stoye J. Proc. of WABI 2006. LNBI, vol. 4175. Heidelberg: Springer Verlag; 2006. A unifying view of genome rearrangements.
-
- Bafna V, Pevzner P. Genome rearrangements and sorting by reversals. In: Proc. of FOCS 1993: 1993. p. 148–57.
-
- Hannenhalli S, Pevzner P. Transforming men into mice (polynomial algorithm for genomic distance problem). In: Proc. of FOCS 1995: 1995. p. 581–92.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

