Analysis and interpretation of transcriptomic data obtained from extended Warburg effect genes in patients with clear cell renal cell carcinoma
- PMID: 25859558
- PMCID: PMC4381708
- DOI: 10.18632/oncoscience.128
Analysis and interpretation of transcriptomic data obtained from extended Warburg effect genes in patients with clear cell renal cell carcinoma
Abstract
Background: Many cancers adopt a metabolism that is characterized by the well-known Warburg effect (aerobic glycolysis). Recently, numerous attempts have been made to treat cancer by targeting one or more gene products involved in this pathway without notable success. This work outlines a transcriptomic approach to identify genes that are highly perturbed in clear cell renal cell carcinoma (CCRCC).
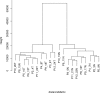
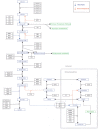
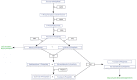
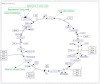
Methods: We developed a model of the extended Warburg effect and outlined the model using Cytoscape. Following this, gene expression fold changes (FCs) for tumor and adjacent normal tissue from patients with CCRCC (GSE6344) were mapped on to the network. Gene expression values with FCs of greater than two were considered as potential targets for treatment of CCRCC.
Results: The Cytoscape network includes glycolysis, gluconeogenesis, the pentose phosphate pathway (PPP), the TCA cycle, the serine/glycine pathway, and partial glutaminolysis and fatty acid synthesis pathways. Gene expression FCs for nine of the 10 CCRCC patients in the GSE6344 data set were consistent with a shift to aerobic glycolysis. Genes involved in glycolysis and the synthesis and transport of lactate were over-expressed, as was the gene that codes for the kinase that inhibits the conversion of pyruvate to acetyl-CoA. Interestingly, genes that code for unique proteins involved in gluconeogenesis were strongly under-expressed as was also the case for the serine/glycine pathway. These latter two results suggest that the role attributed to the M2 isoform of pyruvate kinase (PKM2), frequently the principal isoform of PK present in cancer: i.e. causing a buildup of glucose metabolites that are shunted into branch pathways for synthesis of key biomolecules, may not be operative in CCRCC. The fact that there was no increase in the expression FC of any gene in the PPP is consistent with this hypothesis. Literature protein data generally support the transcriptomic findings.
Conclusions: A number of key genes have been identified that could serve as valid targets for anti-cancer pharmaceutical agents. Genes that are highly over-expressed include ENO2, HK2, PFKP, SLC2A3, PDK1, and SLC16A1. Genes that are highly under-expressed include ALDOB, PKLR, PFKFB2, G6PC, PCK1, FBP1, PC, and SUCLG1.
Keywords: Warburg effect; aerobic glycolysis; clear cell renal cell carcinoma; transcriptomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
The platelet isoform of phosphofructokinase contributes to metabolic reprogramming and maintains cell proliferation in clear cell renal cell carcinoma.Oncotarget. 2016 May 10;7(19):27142-57. doi: 10.18632/oncotarget.8382. Oncotarget. 2016. PMID: 27049827 Free PMC article.
-
PCK1 Regulates Glycolysis and Tumor Progression in Clear Cell Renal Cell Carcinoma Through LDHA.Onco Targets Ther. 2020 Mar 30;13:2613-2627. doi: 10.2147/OTT.S241717. eCollection 2020. Onco Targets Ther. 2020. PMID: 32280238 Free PMC article.
-
Sirtuin 6 and metabolic genes interplay in Warburg effect in cancers.J Clin Biochem Nutr. 2020 May;66(3):169-175. doi: 10.3164/jcbn.19-110. Epub 2020 Feb 14. J Clin Biochem Nutr. 2020. PMID: 32523242 Free PMC article. Review.
-
N-Myc downstream-regulated gene 2 restrains glycolysis and glutaminolysis in clear cell renal cell carcinoma.Oncol Lett. 2017 Dec;14(6):6881-6887. doi: 10.3892/ol.2017.7024. Epub 2017 Sep 22. Oncol Lett. 2017. PMID: 29163707 Free PMC article.
-
Metabolic phenotype of bladder cancer.Cancer Treat Rev. 2016 Apr;45:46-57. doi: 10.1016/j.ctrv.2016.03.005. Epub 2016 Mar 8. Cancer Treat Rev. 2016. PMID: 26975021 Review.
Cited by
-
A 17-Gene Expression Signature for Early Identification of Poor Prognosis in Clear Cell Renal Cell Carcinoma.Cancers (Basel). 2021 Dec 30;14(1):178. doi: 10.3390/cancers14010178. Cancers (Basel). 2021. PMID: 35008342 Free PMC article.
-
Patterns of gene expression characterize T1 and T3 clear cell renal cell carcinoma subtypes.PLoS One. 2019 May 31;14(5):e0216793. doi: 10.1371/journal.pone.0216793. eCollection 2019. PLoS One. 2019. PMID: 31150395 Free PMC article.
-
Long Non-coding RNA LINC01094 Promotes the Development of Clear Cell Renal Cell Carcinoma by Upregulating SLC2A3 via MicroRNA-184.Front Genet. 2020 Sep 23;11:562967. doi: 10.3389/fgene.2020.562967. eCollection 2020. Front Genet. 2020. PMID: 33173535 Free PMC article.
-
Glycolysis-Related Genes Serve as Potential Prognostic Biomarkers in Clear Cell Renal Cell Carcinoma.Oxid Med Cell Longev. 2021 Jan 23;2021:6699808. doi: 10.1155/2021/6699808. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 33564363 Free PMC article.
-
Emerging roles of cytosolic phosphoenolpyruvate kinase 1 (PCK1) in cancer.Biochem Biophys Rep. 2023 Aug 18;35:101528. doi: 10.1016/j.bbrep.2023.101528. eCollection 2023 Sep. Biochem Biophys Rep. 2023. PMID: 37637941 Free PMC article. Review.
References
-
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. - PubMed
-
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
-
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84(6):1014–1020. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

