The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator
- PMID: 25859664
- PMCID: PMC4393102
- DOI: 10.1371/journal.pgen.1005159
The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator
Abstract
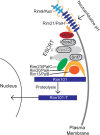
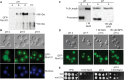
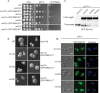
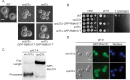
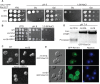
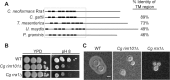
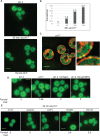
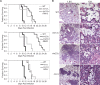
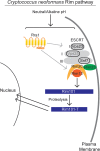
The Rim101/PacC transcription factor acts in a fungal-specific signaling pathway responsible for sensing extracellular pH signals. First characterized in ascomycete fungi such as Aspergillus nidulans and Saccharomyces cerevisiae, the Rim/Pal pathway maintains conserved features among very distantly related fungi, where it coordinates cellular adaptation to alkaline pH signals and micronutrient deprivation. However, it also directs species-specific functions in fungal pathogens such as Cryptococcus neoformans, where it controls surface capsule expression. Moreover, disruption of the Rim pathway central transcription factor, Rim101, results in a strain that causes a hyper-inflammatory response in animal infection models. Using targeted gene deletions, we demonstrate that several genes encoding components of the classical Rim/Pal pathway are present in the C. neoformans genome. Many of these genes are in fact required for Rim101 activation, including members of the ESCRT complex (Vps23 and Snf7), ESCRT-interacting proteins (Rim20 and Rim23), and the predicted Rim13 protease. We demonstrate that in neutral/alkaline pH, Rim23 is recruited to punctate regions on the plasma membrane. This change in Rim23 localization requires upstream ESCRT complex components but does not require other Rim101 proteolysis components, such as Rim20 or Rim13. Using a forward genetics screen, we identified the RRA1 gene encoding a novel membrane protein that is also required for Rim101 protein activation and, like the ESCRT complex, is functionally upstream of Rim23-membrane localization. Homologs of RRA1 are present in other Cryptococcus species as well as other basidiomycetes, but closely related genes are not present in ascomycetes. These findings suggest that major branches of the fungal Kingdom developed different mechanisms to sense and respond to very elemental extracellular signals such as changing pH levels.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Bertuzzi M, Schrettl M, Alcazar-Fuoli L, Cairns TC, Muñoz A, Walker LA, et al. The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014. Oct 16;10(10):e1004413 10.1371/journal.ppat.1004413 - DOI - PMC - PubMed
-
- Huang W, Shang Y, Chen P, Gao Q, Wang C. MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ Microbiol. 2014 Mar 10; - PubMed
-
- Narayan S, Batta K, Colloby P, Tan CY. Cutaneous cryptococcus infection due to C. albidus associated with Sezary syndrome. Br J Dermatol. 2000;143(3):632–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

