Exploiting topic modeling to boost metagenomic reads binning
- PMID: 25859745
- PMCID: PMC4402587
- DOI: 10.1186/1471-2105-16-S5-S2
Exploiting topic modeling to boost metagenomic reads binning
Abstract
Background: With the rapid development of high-throughput technologies, researchers can sequence the whole metagenome of a microbial community sampled directly from the environment. The assignment of these metagenomic reads into different species or taxonomical classes is a vital step for metagenomic analysis, which is referred to as binning of metagenomic data.
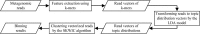
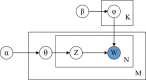
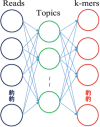
Results: In this paper, we propose a new method TM-MCluster for binning metagenomic reads. First, we represent each metagenomic read as a set of "k-mers" with their frequencies occurring in the read. Then, we employ a probabilistic topic model -- the Latent Dirichlet Allocation (LDA) model to the reads, which generates a number of hidden "topics" such that each read can be represented by a distribution vector of the generated topics. Finally, as in the MCluster method, we apply SKWIC -- a variant of the classical K-means algorithm with automatic feature weighting mechanism to cluster these reads represented by topic distributions.
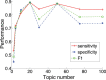
Conclusions: Experiments show that the new method TM-MCluster outperforms major existing methods, including AbundanceBin, MetaCluster 3.0/5.0 and MCluster. This result indicates that the exploitation of topic modeling can effectively improve the binning performance of metagenomic reads.
Figures
Similar articles
-
A New Unsupervised Binning Approach for Metagenomic Sequences Based on N-grams and Automatic Feature Weighting.IEEE/ACM Trans Comput Biol Bioinform. 2014 Jan-Feb;11(1):42-54. doi: 10.1109/TCBB.2013.137. IEEE/ACM Trans Comput Biol Bioinform. 2014. PMID: 26355506
-
MetaCluster-TA: taxonomic annotation for metagenomic data based on assembly-assisted binning.BMC Genomics. 2014;15 Suppl 1(Suppl 1):S12. doi: 10.1186/1471-2164-15-S1-S12. Epub 2014 Jan 24. BMC Genomics. 2014. PMID: 24564377 Free PMC article.
-
Selection of marker genes for genetic barcoding of microorganisms and binning of metagenomic reads by Barcoder software tools.BMC Bioinformatics. 2018 Aug 30;19(1):309. doi: 10.1186/s12859-018-2320-1. BMC Bioinformatics. 2018. PMID: 30165813 Free PMC article.
-
Genome-resolved metagenomics using environmental and clinical samples.Brief Bioinform. 2021 Sep 2;22(5):bbab030. doi: 10.1093/bib/bbab030. Brief Bioinform. 2021. PMID: 33758906 Free PMC article. Review.
-
Classification of metagenomic sequences: methods and challenges.Brief Bioinform. 2012 Nov;13(6):669-81. doi: 10.1093/bib/bbs054. Epub 2012 Sep 8. Brief Bioinform. 2012. PMID: 22962338 Review.
Cited by
-
Decontaminating eukaryotic genome assemblies with machine learning.BMC Bioinformatics. 2017 Dec 1;18(1):533. doi: 10.1186/s12859-017-1941-0. BMC Bioinformatics. 2017. PMID: 29191179 Free PMC article.
-
MetaTopics: an integration tool to analyze microbial community profile by topic model.BMC Genomics. 2017 Jan 25;18(Suppl 1):962. doi: 10.1186/s12864-016-3257-2. BMC Genomics. 2017. PMID: 28198670 Free PMC article.
-
A new method for enhancer prediction based on deep belief network.BMC Bioinformatics. 2017 Oct 16;18(Suppl 12):418. doi: 10.1186/s12859-017-1828-0. BMC Bioinformatics. 2017. PMID: 29072144 Free PMC article.
-
A novel procedure on next generation sequencing data analysis using text mining algorithm.BMC Bioinformatics. 2016 May 13;17(1):213. doi: 10.1186/s12859-016-1075-9. BMC Bioinformatics. 2016. PMID: 27177941 Free PMC article.
-
An overview of topic modeling and its current applications in bioinformatics.Springerplus. 2016 Sep 20;5(1):1608. doi: 10.1186/s40064-016-3252-8. eCollection 2016. Springerplus. 2016. PMID: 27652181 Free PMC article. Review.
References
-
- McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Accurate phylogenetic classification of variable-length dna fragments. Nature Methods. 2006;4(1):63–72. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources