Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility
- PMID: 25860613
- PMCID: PMC4393530
- DOI: 10.1016/j.cell.2015.02.044
Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility
Abstract
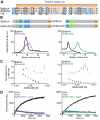
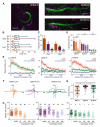
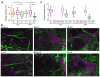
Burkholderia pseudomallei and B. mallei are bacterial pathogens that cause melioidosis and glanders, whereas their close relative B. thailandensis is non-pathogenic. All use the trimeric autotransporter BimA to facilitate actin-based motility, host cell fusion, and dissemination. Here, we show that BimA orthologs mimic different host actin-polymerizing proteins. B. thailandensis BimA activates the host Arp2/3 complex. In contrast, B. pseudomallei and B. mallei BimA mimic host Ena/VASP actin polymerases in their ability to nucleate, elongate, and bundle filaments by associating with barbed ends, as well as in their use of WH2 motifs and oligomerization for activity. Mechanistic differences among BimA orthologs resulted in distinct actin filament organization and motility parameters, which affected the efficiency of cell fusion during infection. Our results identify bacterial Ena/VASP mimics and reveal that pathogens imitate the full spectrum of host actin-polymerizing pathways, suggesting that mimicry of different polymerization mechanisms influences key parameters of infection.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Intracellular bacteria find the right motion.Cell. 2015 Apr 9;161(2):199-200. doi: 10.1016/j.cell.2015.03.035. Cell. 2015. PMID: 25860603
-
Bacterial pathogenesis: Copycat Burkholderia make a move.Nat Rev Microbiol. 2015 Jun;13(6):330. doi: 10.1038/nrmicro3488. Epub 2015 Apr 27. Nat Rev Microbiol. 2015. PMID: 25915638 No abstract available.
References
-
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 1999;274:23549–23557. - PubMed
-
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. - PubMed

