Prokaryotic nucleotide composition is shaped by both phylogeny and the environment
- PMID: 25861819
- PMCID: PMC4453058
- DOI: 10.1093/gbe/evv063
Prokaryotic nucleotide composition is shaped by both phylogeny and the environment
Abstract
The causes of the great variation in nucleotide composition of prokaryotic genomes have long been disputed. Here, we use extensive metagenomic and whole-genome data to demonstrate that both phylogeny and the environment shape prokaryotic nucleotide content. We show that across environments, various phyla are characterized by different mean guanine and cytosine (GC) values as well as by the extent of variation on that mean value. At the same time, we show that GC-content varies greatly as a function of environment, in a manner that cannot be entirely explained by disparities in phylogenetic composition. We find environmentally driven differences in nucleotide content not only between highly diverged environments (e.g., soil, vs. aquatic vs. human gut) but also within a single type of environment. More specifically, we demonstrate that some human guts are associated with a microbiome that is consistently more GC-rich across phyla, whereas others are associated with a more AT-rich microbiome. These differences appear to be driven both by variations in phylogenetic composition and by environmental differences-which are independent of these phylogenetic composition differences. Combined, our results demonstrate that both phylogeny and the environment significantly affect nucleotide composition and that the environmental differences affecting nucleotide composition are far subtler than previously appreciated.
Keywords: GC-content; evolutionary forces; genomic variation; metagenomics; mutation; natural selection.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
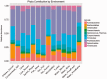

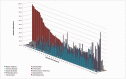
Comment in
-
Highlight-stacking the deck: heritage and environment shape nucleotide composition in prokaryotes.Genome Biol Evol. 2015 May 29;7(5):1390-1. doi: 10.1093/gbe/evv077. Genome Biol Evol. 2015. PMID: 26025642 Free PMC article. No abstract available.
References
-
- Agashe D, Shankar N. The evolution of bacterial DNA base composition. J Exp Zool B Mol Dev Evol. 2014;9999:1–12. - PubMed
-
- Barkovsky EV, Khrustalev VV. Inverse correlation between the GC content of bacterial genomes and their level of preterminal codon usage. Mol Genet Microbiol Virol. 2009;24(1):17–23. - PubMed
-
- Basak S, Ghosh T. On the origin of genomic adaptation at high temperature for prokaryotic organisms. Biochem Biophys Res Commun. 2005;330(3):629–632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

