MetaSV: an accurate and integrative structural-variant caller for next generation sequencing
- PMID: 25861968
- PMCID: PMC4528635
- DOI: 10.1093/bioinformatics/btv204
MetaSV: an accurate and integrative structural-variant caller for next generation sequencing
Abstract
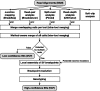
Structural variations (SVs) are large genomic rearrangements that vary significantly in size, making them challenging to detect with the relatively short reads from next-generation sequencing (NGS). Different SV detection methods have been developed; however, each is limited to specific kinds of SVs with varying accuracy and resolution. Previous works have attempted to combine different methods, but they still suffer from poor accuracy particularly for insertions. We propose MetaSV, an integrated SV caller which leverages multiple orthogonal SV signals for high accuracy and resolution. MetaSV proceeds by merging SVs from multiple tools for all types of SVs. It also analyzes soft-clipped reads from alignment to detect insertions accurately since existing tools underestimate insertion SVs. Local assembly in combination with dynamic programming is used to improve breakpoint resolution. Paired-end and coverage information is used to predict SV genotypes. Using simulation and experimental data, we demonstrate the effectiveness of MetaSV across various SV types and sizes.
Availability and implementation: Code in Python is at http://bioinform.github.io/metasv/.
Contact: rd@bina.com
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

