oskar RNA plays multiple noncoding roles to support oogenesis and maintain integrity of the germline/soma distinction
- PMID: 25862242
- PMCID: PMC4436663
- DOI: 10.1261/rna.048298.114
oskar RNA plays multiple noncoding roles to support oogenesis and maintain integrity of the germline/soma distinction
Abstract
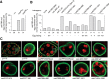
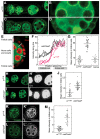
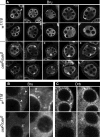
The Drosophila oskar (osk) mRNA is unusual in that it has both coding and noncoding functions. As an mRNA, osk encodes a protein required for embryonic patterning and germ cell formation. Independent of that function, the absence of osk mRNA disrupts formation of the karyosome and blocks progression through oogenesis. Here we show that loss of osk mRNA also affects the distribution of regulatory proteins, relaxing their association with large RNPs within the germline, and allowing them to accumulate in the somatic follicle cells. This and other noncoding functions of the osk mRNA are mediated by multiple sequence elements with distinct roles. One role, provided by numerous binding sites in two distinct regions of the osk 3' UTR, is to sequester the translational regulator Bruno (Bru), which itself controls translation of osk mRNA. This defines a novel regulatory circuit, with Bru restricting the activity of osk, and osk in turn restricting the activity of Bru. Other functional elements, which do not bind Bru and are positioned close to the 3' end of the RNA, act in the oocyte and are essential. Despite the different roles played by the different types of elements contributing to RNA function, mutation of any leads to accumulation of the germline regulatory factors in the follicle cells.
Keywords: Bruno; lncRNA function; oskar; protein sequestration.
© 2015 Kanke et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




Similar articles
-
RNA sequences required for the noncoding function of oskar RNA also mediate regulation of Oskar protein expression by Bicoid Stability Factor.Dev Biol. 2015 Nov 15;407(2):211-23. doi: 10.1016/j.ydbio.2015.09.014. Epub 2015 Oct 9. Dev Biol. 2015. PMID: 26433064 Free PMC article.
-
Different roles for the adjoining and structurally similar A-rich and poly(A) domains of oskar mRNA: Only the A-rich domain is required for oskar noncoding RNA function, which includes MTOC positioning.Dev Biol. 2021 Aug;476:117-127. doi: 10.1016/j.ydbio.2021.03.021. Epub 2021 Mar 31. Dev Biol. 2021. PMID: 33798537 Free PMC article.
-
Region-specific activation of oskar mRNA translation by inhibition of Bruno-mediated repression.PLoS Genet. 2015 Feb 27;11(2):e1004992. doi: 10.1371/journal.pgen.1004992. eCollection 2015. PLoS Genet. 2015. PMID: 25723530 Free PMC article.
-
The mRNA dynamics underpinning translational control mechanisms of Drosophila melanogaster oogenesis.Biochem Soc Trans. 2024 Oct 30;52(5):2087-2099. doi: 10.1042/BST20231293. Biochem Soc Trans. 2024. PMID: 39263986 Free PMC article. Review.
-
The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes.Reproduction. 2009 Apr;137(4):595-617. doi: 10.1530/REP-08-0524. Epub 2009 Feb 18. Reproduction. 2009. PMID: 19225045 Review.
Cited by
-
Peptides encoded by noncoding genes: challenges and perspectives.Signal Transduct Target Ther. 2019 Dec 13;4:57. doi: 10.1038/s41392-019-0092-3. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 31871775 Free PMC article. Review.
-
QTL mapping of natural variation reveals that the developmental regulator bruno reduces tolerance to P-element transposition in the Drosophila female germline.PLoS Biol. 2018 Oct 30;16(10):e2006040. doi: 10.1371/journal.pbio.2006040. eCollection 2018 Oct. PLoS Biol. 2018. PMID: 30376574 Free PMC article.
-
Multiomic analysis of Schistosoma mansoni reveals unique expression profiles in cercarial heads and tails.Commun Biol. 2021 Jul 12;4(1):860. doi: 10.1038/s42003-021-02366-w. Commun Biol. 2021. PMID: 34253841 Free PMC article.
-
Roles for the RNA-Binding Protein Caper in Reproductive Output in Drosophila melanogaster.J Dev Biol. 2022 Dec 23;11(1):2. doi: 10.3390/jdb11010002. J Dev Biol. 2022. PMID: 36648904 Free PMC article.
-
TetraRNA, a tetra-class machine learning model for deciphering the coding potential derivation of RNA world.Comput Struct Biotechnol J. 2025 Mar 26;27:1305-1317. doi: 10.1016/j.csbj.2025.03.039. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40230410 Free PMC article.
References
-
- Bohrmann J, Haas-Assenbaum A 1993. Gap junctions in ovarian follicles of Drosophila melanogaster: inhibition and promotion of dye-coupling between oocyte and follicle cells. Cell Tissue Res 273: 163–173. - PubMed
-
- Cech TR, Steitz JA 2014. The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157: 77–94. - PubMed
-
- Ephrussi A, Dickinson LK, Lehmann R 1991. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
