Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men
- PMID: 25864488
- PMCID: PMC4622564
- DOI: 10.1080/15592294.2015.1022019
Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men
Abstract
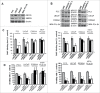
Increasing evidence suggests that aberrant DNA methylation changes may contribute to prostate cancer (PCa) ethnic disparity. To comprehensively identify DNA methylation alterations in PCa disparity, we used the Illumina 450K methylation platform to interrogate the methylation status of 485,577 CpG sites focusing on gene-associated regions of the human genome. Genomic DNA from African-American (AA; 7 normal and 3 cancers) and Caucasian (Cau; 8 normal and 3 cancers) was used in the analysis. Hierarchical clustering analysis identified probe-sets unique to AA and Cau samples, as well as common to both. We selected 25 promoter-associated novel CpG sites most differentially methylated by race (fold change > 1.5-fold; adjusted P < 0.05) and compared the β-value of these sites provided by the Illumina, Inc. array with quantitative methylation obtained by pyrosequencing in 7 prostate cell lines. We found very good concordance of the methylation levels between β-value and pyrosequencing. Gene expression analysis using qRT-PCR in a subset of 8 genes after treatment with 5-aza-2'-deoxycytidine and/or trichostatin showed up-regulation of gene expression in PCa cells. Quantitative analysis of 4 genes, SNRPN, SHANK2, MST1R, and ABCG5, in matched normal and PCa tissues derived from AA and Cau PCa patients demonstrated differential promoter methylation and concomitant differences in mRNA expression in prostate tissues from AA vs. Cau. Regression analysis in normal and PCa tissues as a function of race showed significantly higher methylation prevalence for SNRPN (P = 0.012), MST1R (P = 0.038), and ABCG5 (P < 0.0002) for AA vs. Cau samples. We selected the ABCG5 and SNRPN genes and verified their biological functions by Western blot analysis and siRNA gene knockout effects on cell proliferation and invasion in 4 PCa cell lines (2 AA and 2 Cau patients-derived lines). Knockdown of either ABCG5 or SNRPN resulted in a significant decrease in both invasion and proliferation in Cau PCa cell lines but we did not observe these remarkable loss-of-function effects in AA PCa cell lines. Our study demonstrates how differential genome-wide DNA methylation levels influence gene expression and biological functions in AA and Cau PCa.
Keywords: genome-wide DNA methylation analysis; prostate cancer; pyrosequencing.
Figures





References
-
- Chornokur G, Dalton K, Borysova ME, Kumar NB.. Disparities at presentation, diagnosis, treatment, and survival in African American men, affected by prostate cancer. Prostate 2011; 71:985–97; PMID:21541975; http://dx.doi.org/ 10.1002/pros.21314 - DOI - PMC - PubMed
-
- Kheirandish P, Chinegwundoh F.. Ethnic differences in prostate cancer. Br J Cancer 2011; 105:481–5; PMID:21829203; http://dx.doi.org/ 10.1038/bjc.2011.273 - DOI - PMC - PubMed
-
- Horner R, Ries L, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, et al.. SEER Cancer Statistics Review 1975–2006. National Cancer Institute 2009. www.seer.cancer.gov/archive/csr/1975-2006.
-
- Freedland SJ, Isaacs WB.. Explaining racial differences in prostate cancer in the United States: sociology or biology? Prostate 2005; 62:243–52; PMID:15389726; http://dx.doi.org/ 10.1002/pros.20052 - DOI - PubMed
-
- Hanahan D, Weinberg RA.. The hallmarks of cancer. Cell 2000; 100:57–70; PMID:10647931; http://dx.doi.org/ 10.1016/S0092-8674(00)81683-9 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
