The therapeutic application of CRISPR/Cas9 technologies for HIV
- PMID: 25865334
- PMCID: PMC4581584
- DOI: 10.1517/14712598.2015.1036736
The therapeutic application of CRISPR/Cas9 technologies for HIV
Abstract
Introduction: The use of antiretroviral therapy has led to a significant decrease in morbidity and mortality in HIV-infected individuals. Nevertheless, gene-based therapies represent a promising therapeutic paradigm for HIV-1, as they have the potential for sustained viral inhibition and reduced treatment interventions. One new method amendable to a gene-based therapy is the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein-9 nuclease (Cas9) gene editing system.
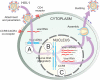
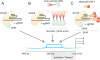
Areas covered: CRISPR/Cas9 can be engineered to successfully modulate an array of disease-causing genetic elements. We discuss the diverse roles that CRISPR/Cas9 may play in targeting HIV and eradicating infection. The Cas9 nuclease coupled with one or more small guide RNAs can target the provirus to mediate excision of the integrated viral genome. Moreover, a modified nuclease-deficient Cas9 fused to transcription activation domains may induce targeted activation of proviral gene expression allowing for the purging of the latent reservoirs. These technologies can also be exploited to target host dependency factors such as the co-receptor CCR5, thus preventing cellular entry of the virus.
Expert opinion: The diversity of the CRISPR/Cas9 technologies offers great promise for targeting different stages of the viral life cycle, and have the capacity for mediating an effective and sustained genetic therapy against HIV.
Keywords: CRISPR/Cas9; HIV-1; RNA therapy; gene activation; gene editing; gene excision; latency.
Figures
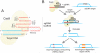


References
-
- Palella FJ, Jr., Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. The New England journal of medicine. 1998 Mar 26;338(13):853–60. - PubMed
-
- Chung J, Scherer LJ, Gu A, Gardner AM, Torres-Coronado M, Epps EW, et al. Optimized lentiviral vectors for HIV gene therapy: multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Molecular therapy : the journal of the American Society of Gene Therapy. 2014 May;22(5):952–63. - PMC - PubMed
-
- Cai M, Yang Y. Targeted genome editing tools for disease modeling and gene therapy. Current gene therapy. 2014 Feb;14(1):2–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
