Bending gradients: how the intestinal stem cell gets its home
- PMID: 25865482
- PMCID: PMC4409931
- DOI: 10.1016/j.cell.2015.03.041
Bending gradients: how the intestinal stem cell gets its home
Abstract
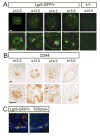
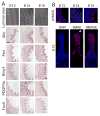
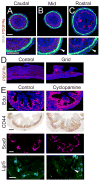
We address the mechanism by which adult intestinal stem cells (ISCs) become localized to the base of each villus during embryonic development. We find that, early in gut development, proliferating progenitors expressing ISC markers are evenly distributed throughout the epithelium, in both the chick and mouse. However, as the villi form, the putative stem cells become restricted to the base of the villi. This shift in the localization is driven by mechanically influenced reciprocal signaling between the epithelium and underlying mesenchyme. Buckling forces physically distort the shape of the morphogenic field, causing local maxima of epithelial signals, in particular Shh, at the tip of each villus. This induces a suite of high-threshold response genes in the underlying mesenchyme to form a signaling center called the "villus cluster." Villus cluster signals, notably Bmp4, feed back on the overlying epithelium to ultimately restrict the stem cells to the base of each villus.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Gradients are shaping up.Cell. 2015 Apr 23;161(3):431-432. doi: 10.1016/j.cell.2015.04.009. Cell. 2015. PMID: 25910201
References
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. - PubMed
-
- Batts LE, Polk DB, Dubois RN, Kulessa H. Bmp signaling is required for intestinal growth and morphogenesis. Dev Dyn. 2006;235:1563–70. - PubMed
-
- Charité J, de Graaff W, Shen S, Deschamps J. Ectopic expression of Hoxb-8 causes duplication of the ZPA in the forelimb and homeotic transformation of axial structures. Cell. 1994;78:589–601. - PubMed
-
- Coulombre AJ, Coulombre JL. Intestinal development, I. Morphogenesis of the villi and musculature. J Embryol Exp Morphol. 1958;6:403–411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

