Lupus Risk Variant Increases pSTAT1 Binding and Decreases ETS1 Expression
- PMID: 25865496
- PMCID: PMC4570281
- DOI: 10.1016/j.ajhg.2015.03.002
Lupus Risk Variant Increases pSTAT1 Binding and Decreases ETS1 Expression
Abstract
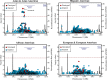
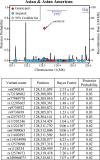
Genetic variants at chromosomal region 11q23.3, near the gene ETS1, have been associated with systemic lupus erythematosus (SLE), or lupus, in independent cohorts of Asian ancestry. Several recent studies have implicated ETS1 as a critical driver of immune cell function and differentiation, and mice deficient in ETS1 develop an SLE-like autoimmunity. We performed a fine-mapping study of 14,551 subjects from multi-ancestral cohorts by starting with genotyped variants and imputing to all common variants spanning ETS1. By constructing genetic models via frequentist and Bayesian association methods, we identified 16 variants that are statistically likely to be causal. We functionally assessed each of these variants on the basis of their likelihood of affecting transcription factor binding, miRNA binding, or chromatin state. Of the four variants that we experimentally examined, only rs6590330 differentially binds lysate from B cells. Using mass spectrometry, we found more binding of the transcription factor signal transducer and activator of transcription 1 (STAT1) to DNA near the risk allele of rs6590330 than near the non-risk allele. Immunoblot analysis and chromatin immunoprecipitation of pSTAT1 in B cells heterozygous for rs6590330 confirmed that the risk allele increased binding to the active form of STAT1. Analysis with expression quantitative trait loci indicated that the risk allele of rs6590330 is associated with decreased ETS1 expression in Han Chinese, but not other ancestral cohorts. We propose a model in which the risk allele of rs6590330 is associated with decreased ETS1 expression and increases SLE risk by enhancing the binding of pSTAT1.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Cui Y., Sheng Y., Zhang X. Genetic susceptibility to SLE: recent progress from GWAS. J. Autoimmun. 2013;41:25–33. - PubMed
-
- Lawrence R.C., Helmick C.G., Arnett F.C., Deyo R.A., Felson D.T., Giannini E.H., Heyse S.P., Hirsch R., Hochberg M.C., Hunder G.G. Estimates of the prevalence of arthritis and selected musculoskeletal disorders in the United States. Arthritis Rheum. 1998;41:778–799. - PubMed
-
- Alarcón-Segovia D., Alarcón-Riquelme M.E., Cardiel M.H., Caeiro F., Massardo L., Villa A.R., Pons-Estel B.A., Grupo Latinoamericano de Estudio del Lupus Eritematoso (GLADEL) Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–1147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K24AR02318/AR/NIAMS NIH HHS/United States
- TR000077/TR/NCATS NIH HHS/United States
- UL1 TR000004/TR/NCATS NIH HHS/United States
- AR057172/AR/NIAMS NIH HHS/United States
- R21AI070304/AI/NIAID NIH HHS/United States
- P60 AR053308/AR/NIAMS NIH HHS/United States
- AR058959/AR/NIAMS NIH HHS/United States
- R01 AI063274/AI/NIAID NIH HHS/United States
- P01AR49084/AR/NIAMS NIH HHS/United States
- U54 GM104938/GM/NIGMS NIH HHS/United States
- AR043814/AR/NIAMS NIH HHS/United States
- P20 RR020143/RR/NCRR NIH HHS/United States
- UL1TR000150/TR/NCATS NIH HHS/United States
- P30 GM103510/GM/NIGMS NIH HHS/United States
- R21 AR065626/AR/NIAMS NIH HHS/United States
- U19AI082714/AI/NIAID NIH HHS/United States
- U19 AI082714/AI/NIAID NIH HHS/United States
- UL1 TR000150/TR/NCATS NIH HHS/United States
- AR051545/AR/NIAMS NIH HHS/United States
- AR060366/AR/NIAMS NIH HHS/United States
- K24AI078004/AI/NIAID NIH HHS/United States
- R56 AI063274/AI/NIAID NIH HHS/United States
- P20 GM103456/GM/NIGMS NIH HHS/United States
- AI083194/AI/NIAID NIH HHS/United States
- AR065626/AR/NIAMS NIH HHS/United States
- P30 AR053483/AR/NIAMS NIH HHS/United States
- AR056360/AR/NIAMS NIH HHS/United States
- P30AR055385/AR/NIAMS NIH HHS/United States
- GM103456/GM/NIGMS NIH HHS/United States
- P60AR064464/AR/NIAMS NIH HHS/United States
- UL1 TR000154/TR/NCATS NIH HHS/United States
- R01 MD007909/MD/NIMHD NIH HHS/United States
- AR053483/AR/NIAMS NIH HHS/United States
- N01 AR062277/AR/NIAMS NIH HHS/United States
- R01 AR056360/AR/NIAMS NIH HHS/United States
- P60 AR064464/AR/NIAMS NIH HHS/United States
- AI063274/AI/NIAID NIH HHS/United States
- HG006828/HG/NHGRI NIH HHS/United States
- AI082714/AI/NIAID NIH HHS/United States
- U01 HG006828/HG/NHGRI NIH HHS/United States
- R01 AR043814/AR/NIAMS NIH HHS/United States
- R37 AI024717/AI/NIAID NIH HHS/United States
- R01 AR042460/AR/NIAMS NIH HHS/United States
- R01 AR057172/AR/NIAMS NIH HHS/United States
- U01 AI101934/AI/NIAID NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- P60AR062755/AR/NIAMS NIH HHS/United States
- S10 RR027015/RR/NCRR NIH HHS/United States
- R01 AR060366/AR/NIAMS NIH HHS/United States
- UL1 RR029882/RR/NCRR NIH HHS/United States
- AR063124/AR/NIAMS NIH HHS/United States
- R01 AR043727/AR/NIAMS NIH HHS/United States
- S10 RR027190/RR/NCRR NIH HHS/United States
- UL1TR000004/TR/NCATS NIH HHS/United States
- 19289/VAC_/Versus Arthritis/United Kingdom
- U01AI101934/AI/NIAID NIH HHS/United States
- P50 AR048940/AR/NIAMS NIH HHS/United States
- P30GM103510/GM/NIGMS NIH HHS/United States
- UL1 TR001425/TR/NCATS NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- S10RR027015/RR/NCRR NIH HHS/United States
- R01 AR063124/AR/NIAMS NIH HHS/United States
- K24 AI078004/AI/NIAID NIH HHS/United States
- R01AR44804/AR/NIAMS NIH HHS/United States
- UL1 TR000077/TR/NCATS NIH HHS/United States
- AI024717/AI/NIAID NIH HHS/United States
- GM104938/GM/NIGMS NIH HHS/United States
- P60AR053308/AR/NIAMS NIH HHS/United States
- U54GM104938/GM/NIGMS NIH HHS/United States
- R21 AI070304/AI/NIAID NIH HHS/United States
- RC2 AR058959/AR/NIAMS NIH HHS/United States
- AR043274/AR/NIAMS NIH HHS/United States
- P01 AI083194/AI/NIAID NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- R01 AR051545/AR/NIAMS NIH HHS/United States
- AR043727/AR/NIAMS NIH HHS/United States
- P30AR053483/AR/NIAMS NIH HHS/United States
- UL1RR029882/RR/NCRR NIH HHS/United States
- R01 AR043274/AR/NIAMS NIH HHS/United States
- P60 AR062755/AR/NIAMS NIH HHS/United States
- U01HG006828/HG/NHGRI NIH HHS/United States
- MD007909/MD/NIMHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

