HOTAIR forms an intricate and modular secondary structure
- PMID: 25866246
- PMCID: PMC4406478
- DOI: 10.1016/j.molcel.2015.03.006
HOTAIR forms an intricate and modular secondary structure
Abstract
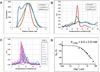
Long noncoding RNAs (lncRNAs) have recently emerged as key players in fundamental cellular processes and diseases, but their functions are poorly understood. HOTAIR is a 2,148-nt-long lncRNA molecule involved in physiological epidermal development and in pathogenic cancer progression, where it has been demonstrated to repress tumor and metastasis suppressor genes. To gain insights into the molecular mechanisms of HOTAIR, we purified it in a stable and homogenous form in vitro, and we determined its functional secondary structure through chemical probing and phylogenetic analysis. The HOTAIR structure reveals a degree of structural organization comparable to well-folded RNAs, like the group II intron, rRNA, or lncRNA steroid receptor activator. It is composed of four independently folding modules, two of which correspond to predicted protein-binding domains. Secondary structure elements that surround protein-binding motifs are evolutionarily conserved. Our work serves as a guide for "navigating" through the lncRNA HOTAIR and ultimately for understanding its function.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

