Resolution improvement by 3D particle averaging in localization microscopy
- PMID: 25866640
- PMCID: PMC4390137
- DOI: 10.1088/2050-6120/3/1/014003
Resolution improvement by 3D particle averaging in localization microscopy
Abstract
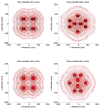
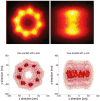
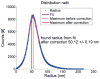
Inspired by recent developments in localization microscopy that applied averaging of identical particles in 2D for increasing the resolution even further, we discuss considerations for alignment (registration) methods for particles in general and for 3D in particular. We detail that traditional techniques for particle registration from cryo electron microscopy based on cross-correlation are not suitable, as the underlying image formation process is fundamentally different. We argue that only localizations, i.e. a set of coordinates with associated uncertainties, are recorded and not a continuous intensity distribution. We present a method that owes to this fact and that is inspired by the field of statistical pattern recognition. In particular we suggest to use an adapted version of the Bhattacharyya distance as a merit function for registration. We evaluate the method in simulations and demonstrate it on three-dimensional super-resolution data of Alexa 647 labelled to the Nup133 protein in the nuclear pore complex of Hela cells. From the simulations we find suggestions that for successful registration the localization uncertainty must be smaller than the distance between labeling sites on a particle. These suggestions are supported by theoretical considerations concerning the attainable resolution in localization microscopy and its scaling behavior as a function of labeling density and localization precision.
Figures






References
-
- Lidke KA, Rieger B, Jovin TM, Heintzmann R. Superresolution by localization of quantum dots using blinking statistics. Optics Express. 2005;13(18):7052–7062. - PubMed
-
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1643–1645. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
