The pbrB gene encodes a laccase required for DHN-melanin synthesis in conidia of Talaromyces (Penicillium) marneffei
- PMID: 25866870
- PMCID: PMC4395095
- DOI: 10.1371/journal.pone.0122728
The pbrB gene encodes a laccase required for DHN-melanin synthesis in conidia of Talaromyces (Penicillium) marneffei
Abstract
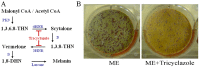
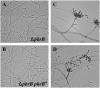
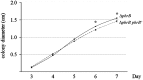
Talaromyces marneffei (Basionym: Penicillium marneffei) is a significant opportunistic fungal pathogen in patients infected with human immunodeficiency virus in Southeast Asia. T. marneffei cells have been shown to become melanized in vivo. Melanins are pigment biopolymers which act as a non-specific protectant against various stressors and which play an important role during virulence in fungi. The synthesis of the two most commonly found melanins in fungi, the eumelanin DOPA-melanin and the allomelanin DHN-melanin, requires the action of laccase enzymes. The T. marneffei genome encodes a number of laccases and this study describes the characterization of one of these, pbrB, during growth and development. A strain carrying a PbrB-GFP fusion shows that pbrB is expressed at high levels during asexual development (conidiation) but not in cells growing vegetatively. The pbrB gene is required for the synthesis of DHN-melanin in conidia and when deleted results in brown pigmented conidia, in contrast to the green conidia of the wild type.
Conflict of interest statement
Figures





Similar articles
-
Talaromyces marneffei laccase modifies THP-1 macrophage responses.Virulence. 2016 Aug 17;7(6):702-17. doi: 10.1080/21505594.2016.1193275. Epub 2016 May 25. Virulence. 2016. PMID: 27224737 Free PMC article.
-
High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei.FEBS J. 2010 Sep;277(18):3750-8. doi: 10.1111/j.1742-4658.2010.07776.x. Epub 2010 Aug 13. FEBS J. 2010. PMID: 20718860
-
Laccases involved in 1,8-dihydroxynaphthalene melanin biosynthesis in Aspergillus fumigatus are regulated by developmental factors and copper homeostasis.Eukaryot Cell. 2013 Dec;12(12):1641-52. doi: 10.1128/EC.00217-13. Epub 2013 Oct 11. Eukaryot Cell. 2013. PMID: 24123270 Free PMC article.
-
Biosynthesis of conidial and sclerotial pigments in Aspergillus species.Appl Microbiol Biotechnol. 2020 Mar;104(6):2277-2286. doi: 10.1007/s00253-020-10347-y. Epub 2020 Jan 23. Appl Microbiol Biotechnol. 2020. PMID: 31974722 Review.
-
Control of morphogenesis in the human fungal pathogen Penicillium marneffei.Int J Med Microbiol. 2002 Oct;292(5-6):331-47. doi: 10.1078/1438-4221-00217. Int J Med Microbiol. 2002. PMID: 12452280 Review.
Cited by
-
Talaromyces marneffei laccase modifies THP-1 macrophage responses.Virulence. 2016 Aug 17;7(6):702-17. doi: 10.1080/21505594.2016.1193275. Epub 2016 May 25. Virulence. 2016. PMID: 27224737 Free PMC article.
-
Role of laccase in the virulence of Talaromyces marneffei: A common link between AIDS-related fungal pathogens?Virulence. 2016 Aug 17;7(6):627-9. doi: 10.1080/21505594.2016.1198867. Epub 2016 Jun 9. Virulence. 2016. PMID: 27282335 Free PMC article. No abstract available.
-
Unraveling the dynamic transcriptomic changes during the dimorphic transition of Talaromyces marneffei through time-course analysis.Front Microbiol. 2024 Apr 24;15:1369349. doi: 10.3389/fmicb.2024.1369349. eCollection 2024. Front Microbiol. 2024. PMID: 38721600 Free PMC article.
-
Characterization of melanin and optimal conditions for pigment production by an endophytic fungus, Spissiomyces endophytica SDBR-CMU319.PLoS One. 2019 Sep 9;14(9):e0222187. doi: 10.1371/journal.pone.0222187. eCollection 2019. PLoS One. 2019. PMID: 31498821 Free PMC article.
-
A Single-Nucleotide Deletion in the Transcription Factor Gene bcsmr1 Causes Sclerotial-Melanogenesis Deficiency in Botrytis cinerea.Front Microbiol. 2017 Dec 12;8:2492. doi: 10.3389/fmicb.2017.02492. eCollection 2017. Front Microbiol. 2017. PMID: 29312200 Free PMC article.
References
-
- Andrianopoulos A. Control of morphogenesis in the human fungal pathogen Penicillium marneffei . Int J Med Microbiol. 2002;292: 331–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

