TAF10 Interacts with the GATA1 Transcription Factor and Controls Mouse Erythropoiesis
- PMID: 25870109
- PMCID: PMC4438247
- DOI: 10.1128/MCB.01370-14
TAF10 Interacts with the GATA1 Transcription Factor and Controls Mouse Erythropoiesis
Abstract
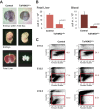
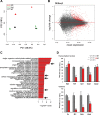
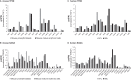
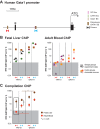
The ordered assembly of a functional preinitiation complex (PIC), composed of general transcription factors (GTFs), is a prerequisite for the transcription of protein-coding genes by RNA polymerase II. TFIID, comprised of the TATA binding protein (TBP) and 13 TBP-associated factors (TAFs), is the GTF that is thought to recognize the promoter sequences allowing site-specific PIC assembly. Transcriptional cofactors, such as SAGA, are also necessary for tightly regulated transcription initiation. The contribution of the two TAF10-containing complexes (TFIID, SAGA) to erythropoiesis remains elusive. By ablating TAF10 specifically in erythroid cells in vivo, we observed a differentiation block accompanied by deregulated GATA1 target genes, including Gata1 itself, suggesting functional cross talk between GATA1 and TAF10. Additionally, we analyzed by mass spectrometry the composition of TFIID and SAGA complexes in mouse and human cells and found that their global integrity is maintained, with minor changes, during erythroid cell differentiation and development. In agreement with our functional data, we show that TAF10 interacts directly with GATA1 and that TAF10 is enriched on the GATA1 locus in human fetal erythroid cells. Thus, our findings demonstrate a cross talk between canonical TFIID and SAGA complexes and cell-specific transcription activators during development and differentiation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures