Phylogenomic analysis and predicted physiological role of the proton-translocating NADH:quinone oxidoreductase (complex I) across bacteria
- PMID: 25873378
- PMCID: PMC4453560
- DOI: 10.1128/mBio.00389-15
Phylogenomic analysis and predicted physiological role of the proton-translocating NADH:quinone oxidoreductase (complex I) across bacteria
Abstract
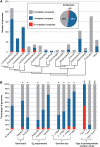
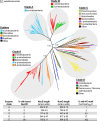
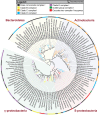
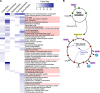
The proton-translocating NADH:quinone oxidoreductase (complex I) is a multisubunit integral membrane enzyme found in the respiratory chains of both bacteria and eukaryotic organelles. Although much research has focused on the enzyme's central role in the mitochondrial respiratory chain, comparatively little is known about its role in the diverse energetic lifestyles of different bacteria. Here, we used a phylogenomic approach to better understand the distribution of complex I across bacteria, the evolution of this enzyme, and its potential roles in shaping the physiology of different bacterial groups. By surveying 970 representative bacterial genomes, we predict complex I to be present in ~50% of bacteria. While this includes bacteria with a wide range of energetic schemes, the presence of complex I is associated with specific lifestyles, including aerobic respiration and specific types of phototrophy (bacteria with only a type II reaction center). A phylogeny of bacterial complex I revealed five main clades of enzymes whose evolution is largely congruent with the evolution of the bacterial groups that encode complex I. A notable exception includes the gammaproteobacteria, whose members encode one of two distantly related complex I enzymes predicted to participate in different types of respiratory chains (aerobic versus anaerobic). Comparative genomic analyses suggest a broad role for complex I in reoxidizing NADH produced from various catabolic reactions, including the tricarboxylic acid (TCA) cycle and fatty acid beta-oxidation. Together, these findings suggest diverse roles for complex I across bacteria and highlight the importance of this enzyme in shaping diverse physiologies across the bacterial domain.
Importance: Living systems use conserved energy currencies, including a proton motive force (PMF), NADH, and ATP. The respiratory chain enzyme, complex I, connects these energy currencies by using NADH produced during nutrient breakdown to generate a PMF, which is subsequently used for ATP synthesis. Our goal is to better understand the role of complex I in bacteria, whose energetic diversity allows us to view its function in a range of biological contexts. We analyzed sequenced bacterial genomes to predict the presence, evolution, and function of complex I in bacteria. We identified five main classes of bacterial complex I and predict that different classes participate in different types of respiratory chains (aerobic and anaerobic). We also predict that complex I helps maintain a cellular redox state by reoxidizing NADH produced from central metabolism. Our findings suggest diverse roles for complex I in bacterial physiology, highlighting the need for future laboratory-based studies.
Copyright © 2015 Spero et al.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

