Changes in expression induced by Epstein-Barr Virus LMP1-CTAR1: potential role of bcl3
- PMID: 25873381
- PMCID: PMC4453583
- DOI: 10.1128/mBio.00441-15
Changes in expression induced by Epstein-Barr Virus LMP1-CTAR1: potential role of bcl3
Abstract
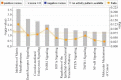
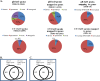
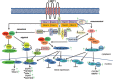
The Epstein-Barr virus protein latent membrane protein 1 (LMP1) has two NF-κB activating domains within its intracellular carboxy terminus (carboxy-terminal activating region 1 [CTAR1] and CTAR2). LMP1-CTAR1 is required for B-lymphocyte transformation, is capable of transforming rodent fibroblasts, and uniquely activates phosphoinositol (PI3) kinase, the noncanonical NF-κB pathway, and expression of the epidermal growth factor receptor (EGFR). In this study, the effects of LMP1-CTAR1 on cellular gene expression were determined by high-throughput sequencing. Additionally, the binding of bcl3 was determined using chromatin immunoprecipitation (ChIP) and sequencing. LMP1-CTAR1 induced few changes in transcription with more genes showing decreased expression. Ingenuity pathway analysis indicated significant enrichment for genes involved in cancer and cellular movement, survival, growth, and proliferation pathways. ChIP in combination with high-throughput sequencing (ChIP-Seq) identified bcl3 binding for more than 2,000 genes in LMP1-CTAR1-expressing cells with more than 90% of the peaks at genes detected within the probable promoter region. Only a small subset of the genes with significant changes in expression had corresponding peaks in the bcl3 ChIP. However, both NFKB2 and PI3 kinase were identified in the bcl3 ChIP. Additionally, many of the predicted upstream regulators for the changes in expression were identified in the bcl3 ChIP. Analysis of the proteins in the NF-κB pathway revealed many changes identified by the high-throughput RNA sequencing (RNA-Seq) and bcl3 ChIP that would likely activate noncanonical NF-κB signaling and possibly inhibit canonical NF-κB signaling. These findings suggest that the two LMP1 signaling domains modulate their combined activity and that the bcl3 transcription factor is likely responsible for some of the unique effects of CTAR1 on cellular expression.
Importance: The Epstein-Barr virus protein latent membrane protein 1 (LMP1) has potent effects on cell growth. LMP1 has two regions, carboxy-terminal activating region 1 (CTAR1) and CTAR2, that distinctly activate NF-κB, a transcription factor complex involved in activation of important host genes. In this study, analysis of the effects on cellular gene expression revealed that CTAR1 significantly affected cellular expression in part through effects on a specific form of NF-κB. The data suggest that LMP1 can activate a distinct subset of host gene expression through its CTAR1 domain which in combination with other signaling effects induced by the CTAR2 domain likely affects cell movement, survival, and growth.
Copyright © 2015 Edwards et al.
Figures
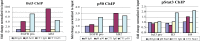

References
-
- Raab-Traub N. 2007. EBV-induced oncogenesis, p 986–1006. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

