Function discovery and structural characterization of a methylphosphonate esterase
- PMID: 25873441
- PMCID: PMC4477287
- DOI: 10.1021/acs.biochem.5b00199
Function discovery and structural characterization of a methylphosphonate esterase
Abstract
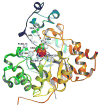
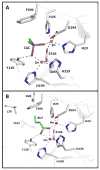
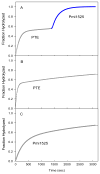
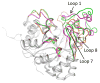
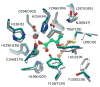
Pmi1525, an enzyme of unknown function from Proteus mirabilis HI4320 and the amidohydrolase superfamily, was cloned, purified to homogeneity, and functionally characterized. The three-dimensional structure of Pmi1525 was determined with zinc and cacodylate bound in the active site (PDB id: 3RHG ). The structure was also determined with manganese and butyrate in the active site (PDB id: 4QSF ). Pmi1525 folds as a distorted (β/α)8-barrel that is typical for members of the amidohydrolase superfamily and cog1735. The substrate profile for Pmi1525 was determined via a strategy that marshaled the utilization of bioinformatics, structural characterization, and focused library screening. The protein was found to efficiently catalyze the hydrolysis of organophosphonate and carboxylate esters. The best substrates identified for Pmi1525 are ethyl 4-nitrophenylmethyl phosphonate (kcat and kcat/Km values of 580 s(-1) and 1.2 × 10(5) M(-1) s(-1), respectively) and 4-nitrophenyl butyrate (kcat and kcat/Km values of 140 s(-1) and 1.4 × 10(5) M(-1) s(-1), respectively). Pmi1525 is stereoselective for the hydrolysis of chiral methylphosphonate esters. The enzyme hydrolyzes the (SP)-enantiomer of isobutyl 4-nitrophenyl methylphosphonate 14 times faster than the corresponding (RP)-enantiomer. The catalytic properties of this enzyme make it an attractive template for the evolution of novel enzymes for the detection, destruction, and detoxification of organophosphonate nerve agents.
Figures





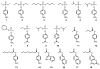
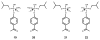
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

