ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase
- PMID: 25873670
- PMCID: PMC4449543
- DOI: 10.1093/jxb/erv138
ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase
Erratum in
-
Corrigendum for: ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels by interacting with arginine decarboxylase.J Exp Bot. 2017 Dec 16;68(21-22):5977. doi: 10.1093/jxb/erv577. J Exp Bot. 2017. PMID: 26856353 Free PMC article. No abstract available.
Abstract
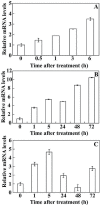
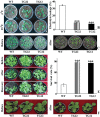
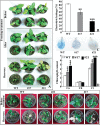
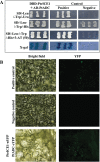
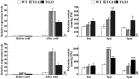
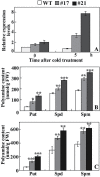
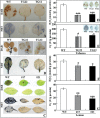
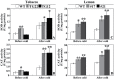
ICE1 (Inducer of CBF Expression 1) encodes a MYC-like basic helix-loop-helix transcription factor that acts as a central regulator of cold response. In this study, we elucidated the function and underlying mechanisms of PtrICE1 from trifoliate orange [Poncirus trifoliata (L.) Raf.]. PtrICE1 was upregulated by cold, dehydration, and salt, with the greatest induction under cold conditions. PtrICE1 was localized in the nucleus and could bind to a MYC-recognizing sequence. Ectopic expression of PtrICE1 in tobacco and lemon conferred enhanced tolerance to cold stresses at either chilling or freezing temperatures. Yeast two-hybrid screening revealed that 21 proteins belonged to the PtrICE1 interactome, in which PtADC (arginine decarboxylase) was confirmed as a bona fide protein interacting with PtrICE1. Transcript levels of ADC genes in the transgenic lines were slightly elevated under normal growth condition but substantially increased under cold conditions, consistent with changes in free polyamine levels. By contrast, accumulation of the reactive oxygen species, H2O2 and O2 (-), was appreciably alleviated in the transgenic lines under cold stress. Higher activities of antioxidant enzymes, such as superoxide dismutase and catalase, were detected in the transgenic lines under cold conditions. Taken together, these results demonstrated that PtrICE1 plays a positive role in cold tolerance, which may be due to modulation of polyamine levels through interacting with the ADC gene.
Keywords: Poncirus trifoliata (L.) Raf.; ROS; bHLH; cold tolerance; polyamine; protein-interacting protein; transcription factor..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



Similar articles
-
Overexpression of a stress-responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis.Plant Physiol Biochem. 2014 May;78:71-9. doi: 10.1016/j.plaphy.2014.02.022. Epub 2014 Mar 5. Plant Physiol Biochem. 2014. PMID: 24636909
-
A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide.Plant Physiol. 2013 Jun;162(2):1178-94. doi: 10.1104/pp.112.210740. Epub 2013 Apr 26. Plant Physiol. 2013. PMID: 23624854 Free PMC article.
-
An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis.J Exp Bot. 2011 May;62(8):2899-914. doi: 10.1093/jxb/erq463. Epub 2011 Jan 31. J Exp Bot. 2011. PMID: 21282323
-
Transcriptional regulation of bHLH during plant response to stress.Biochem Biophys Res Commun. 2018 Sep 5;503(2):397-401. doi: 10.1016/j.bbrc.2018.07.123. Epub 2018 Jul 27. Biochem Biophys Res Commun. 2018. PMID: 30057319 Review.
-
Polyamines: molecules with regulatory functions in plant abiotic stress tolerance.Planta. 2010 May;231(6):1237-49. doi: 10.1007/s00425-010-1130-0. Epub 2010 Mar 11. Planta. 2010. PMID: 20221631 Review.
Cited by
-
A genetic genomics-expression approach reveals components of the molecular mechanisms beyond the cell wall that underlie peach fruit woolliness due to cold storage.Plant Mol Biol. 2016 Nov;92(4-5):483-503. doi: 10.1007/s11103-016-0526-z. Epub 2016 Oct 6. Plant Mol Biol. 2016. PMID: 27714490
-
ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1.Sci Rep. 2015 Dec 2;5:17620. doi: 10.1038/srep17620. Sci Rep. 2015. PMID: 26626798 Free PMC article.
-
Responses of Manila Grass (Zoysia matrella) to chilling stress: From transcriptomics to physiology.PLoS One. 2020 Jul 20;15(7):e0235972. doi: 10.1371/journal.pone.0235972. eCollection 2020. PLoS One. 2020. PMID: 32687533 Free PMC article.
-
Alfalfa (Medicago sativa L.) MsCML46 gene encoding calmodulin-like protein confers tolerance to abiotic stress in tobacco.Plant Cell Rep. 2021 Oct;40(10):1907-1922. doi: 10.1007/s00299-021-02757-7. Epub 2021 Jul 28. Plant Cell Rep. 2021. PMID: 34322731
-
A Novel NAC Transcription Factor, PbeNAC1, of Pyrus betulifolia Confers Cold and Drought Tolerance via Interacting with PbeDREBs and Activating the Expression of Stress-Responsive Genes.Front Plant Sci. 2017 Jun 30;8:1049. doi: 10.3389/fpls.2017.01049. eCollection 2017. Front Plant Sci. 2017. PMID: 28713394 Free PMC article.
References
-
- Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF. 2010. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249. - PubMed
-
- Alcázar R, Cuevas JC, Planas J, Zarza X, Bortolotti C, Carrasco P, Salinas J, Tiburcio AF, Altabella T. 2011. Integration of polyamines in the cold acclimation response. Plant Science 180, 31–38. - PubMed
-
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. 2003. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

