Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy
- PMID: 25873913
- PMCID: PMC4379921
- DOI: 10.3389/fmicb.2015.00242
Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy
Abstract
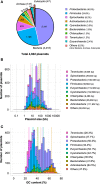
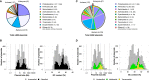
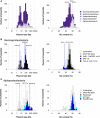
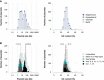
Plasmids are important "vehicles" for the communication of genetic information between bacteria. The exchange of plasmids transmits pathogenically and environmentally relevant traits to the host bacteria, promoting their rapid evolution and adaptation to various environments. Over the past six decades, a large number of plasmids have been identified and isolated from different microbes. With the revolution of sequencing technology, more than 4600 complete sequences of plasmids found in bacteria, archaea, and eukaryotes have been determined. The classification of a wide variety of plasmids is not only important to understand their features, host ranges, and microbial evolution but is also necessary to effectively use them as genetic tools for microbial engineering. This review summarizes the current situation of the classification of fully sequenced plasmids based on their host taxonomy and their features of replication and conjugative transfer. The majority of the fully sequenced plasmids are found in bacteria in the Proteobacteria, Firmicutes, Spirochaetes, Actinobacteria, Cyanobacteria and Euryarcheota phyla, and key features of each phylum are included. Recent advances in the identification of novel types of plasmids and plasmid transfer by culture-independent methods using samples from natural environments are also discussed.
Keywords: Inc group; conjugative transfer; host; plasmid; replication.
Figures
References
-
- Adamczyk M., Jagura-Burdzy G. (2003). Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50, 425–453. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

