Integrated analysis reveals microRNA networks coordinately expressed with key proteins in breast cancer
- PMID: 25873999
- PMCID: PMC4396592
- DOI: 10.1186/s13073-015-0135-5
Integrated analysis reveals microRNA networks coordinately expressed with key proteins in breast cancer
Abstract
Background: The role played by microRNAs in the deregulation of protein expression in breast cancer is only partly understood. To gain insight, the combined effect of microRNA and mRNA expression on protein expression was investigated in three independent data sets.
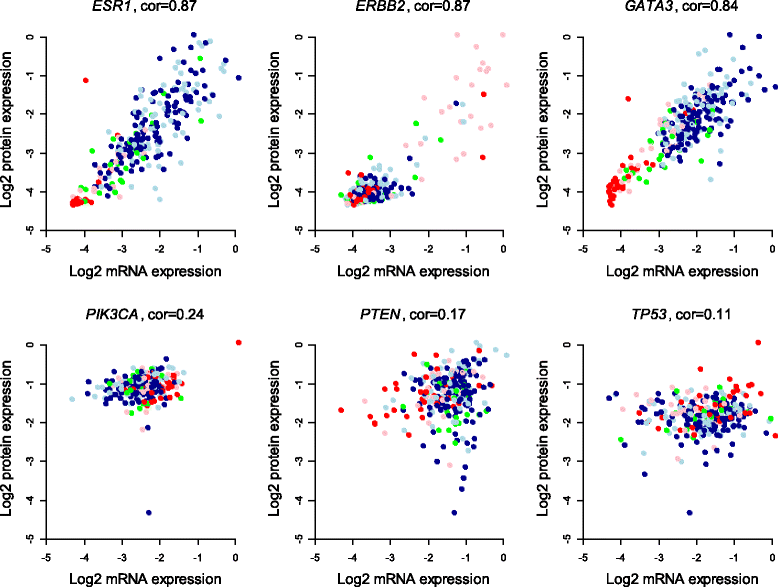
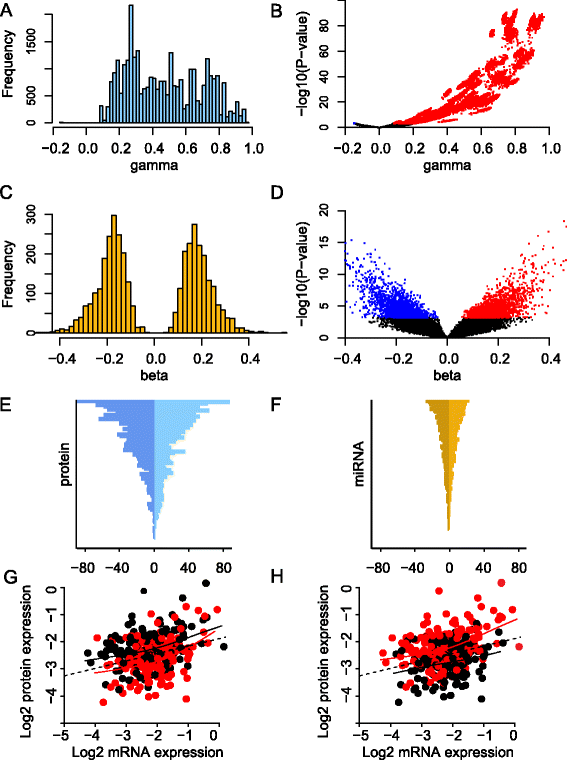
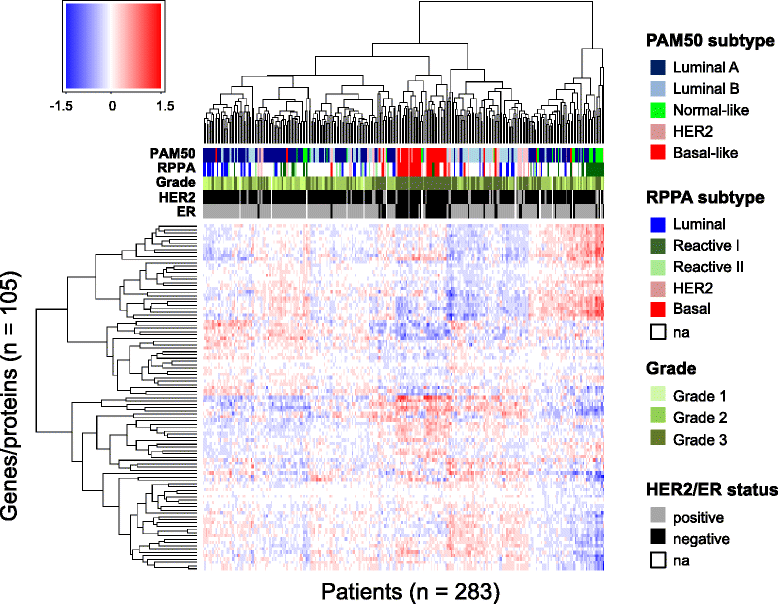
Methods: Protein expression was modeled as a multilinear function of powers of mRNA and microRNA expression. The model was first applied to mRNA and protein expression for 105 selected cancer-associated genes and to genome-wide microRNA expression from 283 breast tumors. The model considered both the effect of one microRNA at a time and all microRNAs combined. In the latter case the Lasso penalized regression method was applied to detect the simultaneous effect of multiple microRNAs.
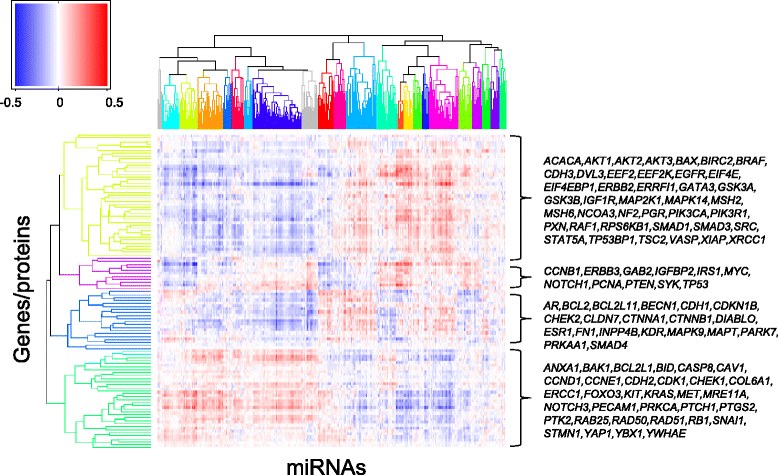
Results: An interactome map for breast cancer representing all direct and indirect associations between the expression of microRNAs and proteins was derived. A pattern of extensive coordination between microRNA and protein expression in breast cancer emerges, with multiple clusters of microRNAs being associated with multiple clusters of proteins. Results were subsequently validated in two independent breast cancer data sets. A number of the microRNA-protein associations were functionally validated in a breast cancer cell line.
Conclusions: A comprehensive map is derived for the co-expression in breast cancer of microRNAs and 105 proteins with known roles in cancer, after filtering out the in-cis effect of mRNA expression. The analysis suggests that group action by several microRNAs to deregulate the expression of proteins is a common modus operandi in breast cancer.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

