Bistability: requirements on cell-volume, protein diffusion, and thermodynamics
- PMID: 25874711
- PMCID: PMC4398428
- DOI: 10.1371/journal.pone.0121681
Bistability: requirements on cell-volume, protein diffusion, and thermodynamics
Abstract
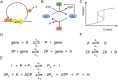
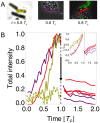
Bistability is considered wide-spread among bacteria and eukaryotic cells, useful, e.g., for enzyme induction, bet hedging, and epigenetic switching. However, this phenomenon has mostly been described with deterministic dynamic or well-mixed stochastic models. Here, we map known biological bistable systems onto the well-characterized biochemical Schlögl model, using analytical calculations and stochastic spatiotemporal simulations. In addition to network architecture and strong thermodynamic driving away from equilibrium, we show that bistability requires fine-tuning towards small cell volumes (or compartments) and fast protein diffusion (well mixing). Bistability is thus fragile and hence may be restricted to small bacteria and eukaryotic nuclei, with switching triggered by volume changes during the cell cycle. For large volumes, single cells generally loose their ability for bistable switching and instead undergo a first-order phase transition.
Conflict of interest statement
Figures






Similar articles
-
Stochastic dynamics and non-equilibrium thermodynamics of a bistable chemical system: the Schlögl model revisited.J R Soc Interface. 2009 Oct 6;6(39):925-40. doi: 10.1098/rsif.2008.0476. Epub 2008 Dec 18. J R Soc Interface. 2009. PMID: 19095615 Free PMC article.
-
Confinement and diffusion modulate bistability and stochastic switching in a reaction network with positive feedback.J Chem Phys. 2016 Jan 7;144(1):015102. doi: 10.1063/1.4939219. J Chem Phys. 2016. PMID: 26747820
-
Stochastic models for regulatory networks of the genetic toggle switch.Proc Natl Acad Sci U S A. 2006 May 30;103(22):8372-7. doi: 10.1073/pnas.0507818103. Epub 2006 May 19. Proc Natl Acad Sci U S A. 2006. PMID: 16714385 Free PMC article.
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
-
Bistability, epigenetics, and bet-hedging in bacteria.Annu Rev Microbiol. 2008;62:193-210. doi: 10.1146/annurev.micro.62.081307.163002. Annu Rev Microbiol. 2008. PMID: 18537474 Review.
Cited by
-
Entropy production selects nonequilibrium states in multistable systems.Sci Rep. 2017 Oct 31;7(1):14437. doi: 10.1038/s41598-017-14485-8. Sci Rep. 2017. PMID: 29089531 Free PMC article.
-
Quantifying the roles of space and stochasticity in computer simulations for cell biology and cellular biochemistry.Mol Biol Cell. 2021 Jan 15;32(2):186-210. doi: 10.1091/mbc.E20-08-0530. Epub 2020 Nov 25. Mol Biol Cell. 2021. PMID: 33237849 Free PMC article.
-
A minimal model of burst-noise induced bistability.PLoS One. 2017 Apr 27;12(4):e0176410. doi: 10.1371/journal.pone.0176410. eCollection 2017. PLoS One. 2017. PMID: 28448638 Free PMC article.
-
Effects of cell cycle noise on excitable gene circuits.Phys Biol. 2016 Nov 30;13(6):066007. doi: 10.1088/1478-3975/13/6/066007. Phys Biol. 2016. PMID: 27902489 Free PMC article.
-
Decorating the surface of Escherichia coli with bacterial lipoproteins: a comparative analysis of different display systems.Microb Cell Fact. 2021 Feb 2;20(1):33. doi: 10.1186/s12934-021-01528-z. Microb Cell Fact. 2021. PMID: 33531008 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

