Preferential targeting of Nav1.6 voltage-gated Na+ Channels to the axon initial segment during development
- PMID: 25874799
- PMCID: PMC4398423
- DOI: 10.1371/journal.pone.0124397
Preferential targeting of Nav1.6 voltage-gated Na+ Channels to the axon initial segment during development
Abstract
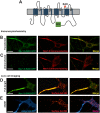
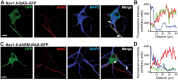
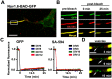
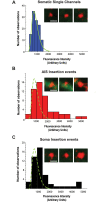
During axonal maturation, voltage-gated sodium (Nav) channels accumulate at the axon initial segment (AIS) at high concentrations. This localization is necessary for the efficient initiation of action potentials. The mechanisms underlying channel trafficking to the AIS during axonal development have remained elusive due to a lack of Nav reagents suitable for high resolution imaging of channels located specifically on the cell surface. Using an optical pulse-chase approach in combination with a novel Nav1.6 construct containing an extracellular biotinylation domain we demonstrate that Nav1.6 channels are preferentially inserted into the AIS membrane during neuronal development via direct vesicular trafficking. Single-molecule tracking illustrates that axonal channels are immediately immobilized following delivery, while channels delivered to the soma are often mobile. Neither a Nav1.6 channel lacking the ankyrin-binding motif nor a chimeric Kv2.1 channel containing the Nav ankyrinG-binding domain show preferential AIS insertion. Together these data support a model where ankyrinG-binding is required for preferential Nav1.6 insertion into the AIS plasma membrane. In contrast, ankyrinG-binding alone does not confer the preferential delivery of proteins to the AIS.
Conflict of interest statement
Figures



References
-
- Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, et al. Primary Structure and Functional Expression of the Beta 1 Subunit of the Rat Brain Sodium Channel. Science. 1992. May 8;256(5058):839–42. . - PubMed
-
- Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. Xlvii. Nomenclature and Structure-Function Relationships of Voltage-Gated Sodium Channels. Pharmacol Rev. 2005. December;57(4):397–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

