Visualizing the indefinable: three-dimensional complexity of 'infectious diseases'
- PMID: 25875169
- PMCID: PMC4397090
- DOI: 10.1371/journal.pone.0123674
Visualizing the indefinable: three-dimensional complexity of 'infectious diseases'
Abstract
Background: The words 'infection' and 'inflammation' lack specific definitions. Here, such words are not defined. Instead, the ability to visualize host-microbial interactions was explored.
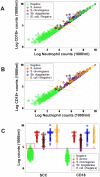
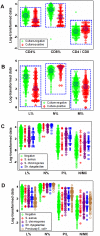
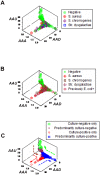
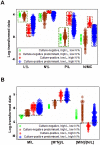
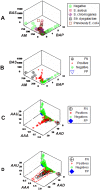
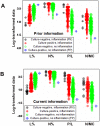
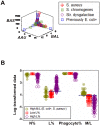
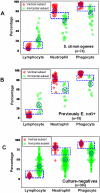
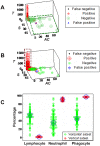
Methods: Leukocyte differential counts and four bacterial species (Staphylococcus aureus, Streptococcus dysgalactiae, Staphylococcus chromogenes, and Escherichia coli) were determined or isolated in a cross-sectional and randomized study conducted with 611 bovine milk samples. Two paradigms were evaluated: (i) the classic one, which measures non-structured (count or percent) data; and (ii) a method that, using complex data structures, detects and differentiates three-dimensional (3D) interactions among lymphocytes (L), macrophages (M), and neutrophils (N).
Results: Classic analyses failed to differentiate bacterial-positive (B+) from -negative (B-) observations: B- and B+ data overlapped, even when statistical significance was achieved. In contrast, the alternative approach showed distinct patterns, such as perpendicular data inflections, which discriminated microbial-negative/mononuclear cell-predominating (MCP) from microbial-positive/phagocyte-predominating (PP) subsets. Two PP subcategories were distinguished, as well as PP/culture-negative (false-negative) and MCP/culture-positive (false-positive) observations. In 3D space, MCP and PP subsets were perpendicular to one another, displaying ≥ 91% specificity or sensitivity. Findings supported five inferences: (i) disease is not always ruled out by negative bacterial tests; (ii) low total cell counts can coexist with high phagocyte percents; (iii) neither positive bacterial isolation nor high cell counts always coincide with PP profiles; (iv) statistical significance is not synonymous with discrimination; and (v) hidden relationships cannot be detected when simple (non-structured) data formats are used and statistical analyses are performed before data subsets are identified, but can be uncovered when complexity is investigated.
Conclusions: Pattern recognition-based assessments can detect host-microbial interactions usually unobserved. Such cutoff-free, confidence interval-free, gold standard-free approaches provide interpretable information on complex entities, such as 'infection' and 'inflammation', even without definitions. To investigate disease dynamics, combinations of observational and experimental longitudinal studies, on human and non-human infections, are recommended.
Conflict of interest statement
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

