Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism
- PMID: 25875217
- PMCID: PMC4626538
- DOI: 10.1007/s10545-015-9843-7
Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism
Erratum in
-
Erratum to: Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism.J Inherit Metab Dis. 2016 Sep;39(5):757. doi: 10.1007/s10545-016-9944-y. J Inherit Metab Dis. 2016. PMID: 27225280 Free PMC article. No abstract available.
Abstract
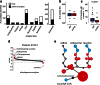
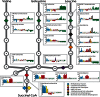
Global metabolic profiling currently achievable by untargeted mass spectrometry-based metabolomic platforms has great potential to advance our understanding of human disease states, including potential utility in the detection of novel and known inborn errors of metabolism (IEMs). There are few studies of the technical reproducibility, data analysis methods, and overall diagnostic capabilities when this technology is applied to clinical specimens for the diagnosis of IEMs. We explored the clinical utility of a metabolomic workflow capable of routinely generating semi-quantitative z-score values for ~900 unique compounds, including ~500 named human analytes, in a single analysis of human plasma. We tested the technical reproducibility of this platform and applied it to the retrospective diagnosis of 190 individual plasma samples, 120 of which were collected from patients with a confirmed IEM. Our results demonstrate high intra-assay precision and linear detection for the majority compounds tested. Individual metabolomic profiles provided excellent sensitivity and specificity for the detection of a wide range of metabolic disorders and identified novel biomarkers for some diseases. With this platform, it is possible to use one test to screen for dozens of IEMs that might otherwise require ordering multiple unique biochemical tests. However, this test may yield false negative results for certain disorders that would be detected by a more well-established quantitative test and in its current state should be considered a supplementary test. Our findings describe a novel approach to metabolomic analysis of clinical specimens and demonstrate the clinical utility of this technology for prospective screening of IEMs.
Figures


References
-
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81(16):6656–6667. doi: 10.1021/ac901536h. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

