Unraveling the molecular signatures of oxidative phosphorylation to cope with the nutritionally changing metabolic capabilities of liver and muscle tissues in farmed fish
- PMID: 25875231
- PMCID: PMC4398389
- DOI: 10.1371/journal.pone.0122889
Unraveling the molecular signatures of oxidative phosphorylation to cope with the nutritionally changing metabolic capabilities of liver and muscle tissues in farmed fish
Abstract
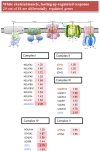
Mitochondrial oxidative phosphorylation provides over 90% of the energy produced by aerobic organisms, therefore the regulation of mitochondrial activity is a major issue for coping with the changing environment and energy needs. In fish, there is a large body of evidence of adaptive changes in enzymatic activities of the OXPHOS pathway, but less is known at the transcriptional level and the first aim of the present study was to define the molecular identity of the actively transcribed subunits of the mitochondrial respiratory chain of a livestock animal, using gilthead sea bream as a model of farmed fish with a high added value for European aquaculture. Extensive BLAST searches in our transcriptomic database (www.nutrigroup-iats.org/seabreamdb) yielded 97 new sequences with a high coverage of catalytic, regulatory and assembly factors of Complex I to V. This was the basis for the development of a PCR array for the simultaneous profiling of 88 selected genes. This new genomic resource allowed the differential gene expression of liver and muscle tissues in a model of 10 fasting days. A consistent down-regulated response involving 72 genes was made by the liver, whereas an up-regulated response with 29 and 10 differentially expressed genes was found in white skeletal muscle and heart, respectively. This differential regulation was mostly mediated by nuclear-encoded genes (skeletal muscle) or both mitochondrial- and nuclear-encoded genes (liver, heart), which is indicative of a complex and differential regulation of mitochondrial and nuclear genomes, according to the changes in the lipogenic activity of liver and the oxidative capacity of glycolytic and highly oxidative muscle tissues. These insights contribute to the identification of the most responsive elements of OXPHOS in each tissue, which is of relevance for the appropriate gene targeting of nutritional and/or environmental metabolic disturbances in livestock animals.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

