Genetics, genomics and evolution of ergot alkaloid diversity
- PMID: 25875294
- PMCID: PMC4417967
- DOI: 10.3390/toxins7041273
Genetics, genomics and evolution of ergot alkaloid diversity
Abstract
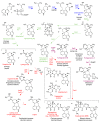
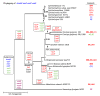
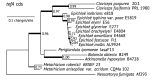
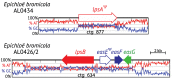
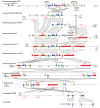
The ergot alkaloid biosynthesis system has become an excellent model to study evolutionary diversification of specialized (secondary) metabolites. This is a very diverse class of alkaloids with various neurotropic activities, produced by fungi in several orders of the phylum Ascomycota, including plant pathogens and protective plant symbionts in the family Clavicipitaceae. Results of comparative genomics and phylogenomic analyses reveal multiple examples of three evolutionary processes that have generated ergot-alkaloid diversity: gene gains, gene losses, and gene sequence changes that have led to altered substrates or product specificities of the enzymes that they encode (neofunctionalization). The chromosome ends appear to be particularly effective engines for gene gains, losses and rearrangements, but not necessarily for neofunctionalization. Changes in gene expression could lead to accumulation of various pathway intermediates and affect levels of different ergot alkaloids. Genetic alterations associated with interspecific hybrids of Epichloë species suggest that such variation is also selectively favored. The huge structural diversity of ergot alkaloids probably represents adaptations to a wide variety of ecological situations by affecting the biological spectra and mechanisms of defense against herbivores, as evidenced by the diverse pharmacological effects of ergot alkaloids used in medicine.
Figures





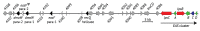
References
-
- Billings M. The Crusades: Five Centuries of Holy Wars. Sterling Publishing Company; New York, NY, USA: 1996.
-
- Zadoks J.C. On the Political Economy of Plant Disease Epidemics: Capita Selecta in Historical Epidemiology. Wageningen Academic Publishers; Wageningen, The Netherlands: 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

