Construction of an ortholog database using the semantic web technology for integrative analysis of genomic data
- PMID: 25875762
- PMCID: PMC4395280
- DOI: 10.1371/journal.pone.0122802
Construction of an ortholog database using the semantic web technology for integrative analysis of genomic data
Abstract
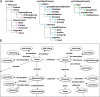
Recently, various types of biological data, including genomic sequences, have been rapidly accumulating. To discover biological knowledge from such growing heterogeneous data, a flexible framework for data integration is necessary. Ortholog information is a central resource for interlinking corresponding genes among different organisms, and the Semantic Web provides a key technology for the flexible integration of heterogeneous data. We have constructed an ortholog database using the Semantic Web technology, aiming at the integration of numerous genomic data and various types of biological information. To formalize the structure of the ortholog information in the Semantic Web, we have constructed the Ortholog Ontology (OrthO). While the OrthO is a compact ontology for general use, it is designed to be extended to the description of database-specific concepts. On the basis of OrthO, we described the ortholog information from our Microbial Genome Database for Comparative Analysis (MBGD) in the form of Resource Description Framework (RDF) and made it available through the SPARQL endpoint, which accepts arbitrary queries specified by users. In this framework based on the OrthO, the biological data of different organisms can be integrated using the ortholog information as a hub. Besides, the ortholog information from different data sources can be compared with each other using the OrthO as a shared ontology. Here we show some examples demonstrating that the ortholog information described in RDF can be used to link various biological data such as taxonomy information and Gene Ontology. Thus, the ortholog database using the Semantic Web technology can contribute to biological knowledge discovery through integrative data analysis.
Conflict of interest statement
Figures




References
-
- Fitch WM. Distinguishing homologous from analogous proteins. Systematic zoology. 1970;19: 99–113. - PubMed
-
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annual review of genetics. 2005;39: 309–38. - PubMed
-
- Berners-Lee T, Hendler J. Publishing on the semantic web. Nature. 2001;410: 1023–4. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

