Statistical distance as a measure of physiological dysregulation is largely robust to variation in its biomarker composition
- PMID: 25875923
- PMCID: PMC4395377
- DOI: 10.1371/journal.pone.0122541
Statistical distance as a measure of physiological dysregulation is largely robust to variation in its biomarker composition
Abstract
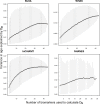
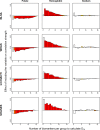
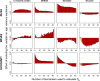
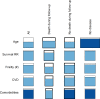
Physiological dysregulation may underlie aging and many chronic diseases, but is challenging to quantify because of the complexity of the underlying systems. Recently, we described a measure of physiological dysregulation, DM, that uses statistical distance to assess the degree to which an individual's biomarker profile is normal versus aberrant. However, the sensitivity of DM to details of the calculation method has not yet been systematically assessed. In particular, the number and choice of biomarkers and the definition of the reference population (RP, the population used to define a "normal" profile) may be important. Here, we address this question by validating the method on 44 common clinical biomarkers from three longitudinal cohort studies and one cross-sectional survey. DMs calculated on different biomarker subsets show that while the signal of physiological dysregulation increases with the number of biomarkers included, the value of additional markers diminishes as more are added and inclusion of 10-15 is generally sufficient. As long as enough markers are included, individual markers have little effect on the final metric, and even DMs calculated from mutually exclusive groups of markers correlate with each other at r~0.4-0.5. We also used data subsets to generate thousands of combinations of study populations and RPs to address sensitivity to differences in age range, sex, race, data set, sample size, and their interactions. Results were largely consistent (but not identical) regardless of the choice of RP; however, the signal was generally clearer with a younger and healthier RP, and RPs too different from the study population performed poorly. Accordingly, biomarker and RP choice are not particularly important in most cases, but caution should be used across very different populations or for fine-scale analyses. Biologically, the lack of sensitivity to marker choice and better performance of younger, healthier RPs confirm an interpretation of DM physiological dysregulation and as an emergent property of a complex system.
Conflict of interest statement
Figures



References
-
- Kirkwood TBL. Understanding the Odd Science of Aging. Cell. 2005; 120: 437–447. - PubMed
-
- Medvedev ZA. An attempt at a rational classification of theories of ageing. Biological Reviews. 1990; 65: 375–398. - PubMed
-
- Cohen AA, Martin LB, Wingfield JC, McWilliams SR, Dunne JA. Physiological regulatory networks: ecological roles and evolutionary constraints. Trends in Ecology & Evolution. 2012; 27: 428–435. - PubMed
-
- Ferrucci L, Windham BG, Fried LP. Frailty in older persons. Genus. 2005; 61: 39–53.
-
- Fried LP, Hadley EC, Walston JD, Newman AB, Guralnik JM, Studenski S, et al. From Bedside to Bench: Research Agenda for Frailty. Sci Aging Knowl Environ. 2005;2005: pe24-. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

