Sinonasal microbiome sampling: a comparison of techniques
- PMID: 25876035
- PMCID: PMC4396979
- DOI: 10.1371/journal.pone.0123216
Sinonasal microbiome sampling: a comparison of techniques
Abstract
Background: The role of the sino-nasal microbiome in CRS remains unclear. We hypothesized that the bacteria within mucosal-associated biofilms may be different from the more superficial-lying, free-floating bacteria in the sinuses and that this may impact on the microbiome results obtained. This study investigates whether there is a significant difference in the microbiota of a sinonasal mucosal tissue sample versus a swab sample.
Methods: Cross-sectional study with paired design. Mucosal biopsy and swab samples were obtained intra-operatively from the ethmoid sinuses of 6 patients with CRS. Extracted DNA was sequenced on a Roche-454 sequencer using 16S-rRNA gene targeted primers. Data were analyzed using QIIME 1.8 software package.
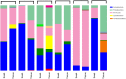
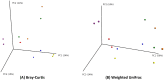
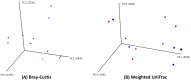
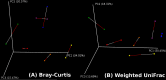
Results: At a maximum subsampling depth of 1,100 reads, the mean observed species richness was 33.3 species (30.6 for swab, versus 36 for mucosa; p > 0.05). There was no significant difference in phylogenetic and non-phylogenetic alpha diversity metrics (Faith's PD_Whole_Tree and Shannon's index) between the two sampling methods (p > 0.05). The type of sample also had no significant effect on phylogenetic and non-phylogenetic beta diversity metrics (Unifrac and Bray-Curtis; p > 0.05).
Conclusion: We observed no significant difference between the microbiota of mucosal tissue and swab samples. This suggests that less invasive swab samples are representative of the sinonasal mucosa microbiome and can be used for future sinonasal microbiome studies.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

